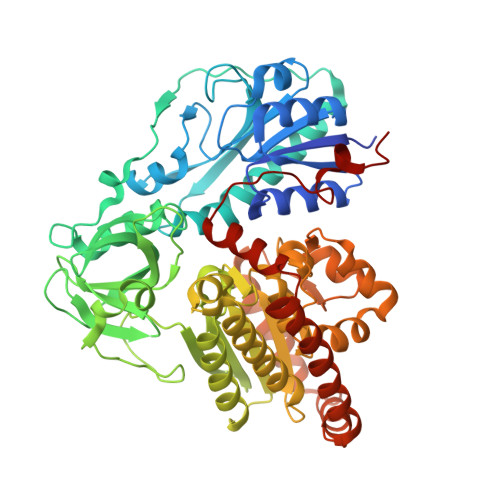
Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Muraki, N., Ishii, K., Uchiyama, S., Itoh, S.G., Okumura, H., Aono, S.(2019) Commun Biol 2: 385-385
- PubMed: 31646188
- DOI: https://doi.org/10.1038/s42003-019-0631-z
- Primary Citation of Related Structures:
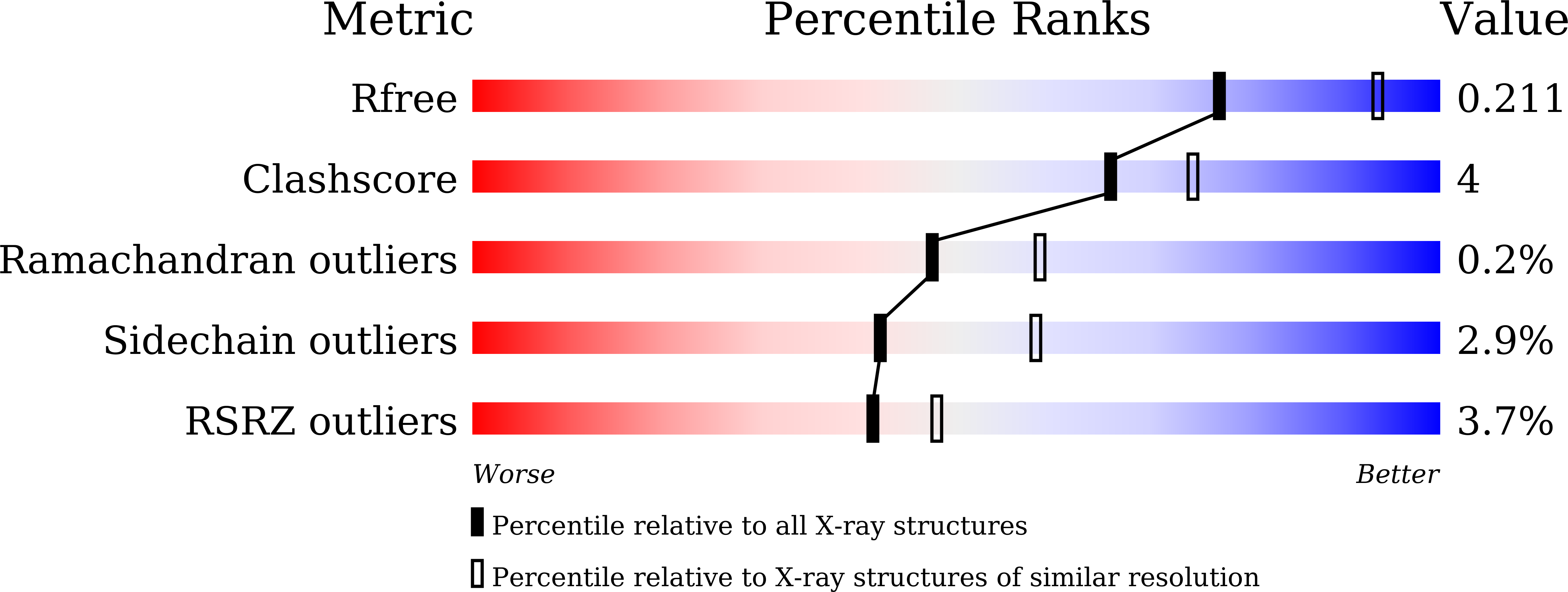
6J0P, 6J1E, 6J1F, 6J1G, 6J1H, 6J1I, 6J1J - PubMed Abstract:
Several accessory proteins are required for the assembly of the metal centers in hydrogenases. In NiFe-hydrogenases, CO and CN - are coordinated to the Fe in the NiFe dinuclear cluster of the active center. Though these diatomic ligands are biosynthesized enzymatically, detail mechanisms of their biosynthesis remain unclear. Here, we report the structural characterization of HypX responsible for CO biosynthesis to assemble the active site of NiFe hydrogenase. CoA is constitutionally bound in HypX. Structural characterization of HypX suggests that the formyl-group transfer will take place from N 10 -formyl-THF to CoA to form formyl-CoA in the N-terminal domain of HypX, followed by decarbonylation of formyl-CoA to produce CO in the C-terminal domain though the direct experimental results are not available yet. The conformation of CoA accommodated in the continuous cavity connecting the N- and C-terminal domains will interconvert between the extended and the folded conformations for HypX catalysis.
Organizational Affiliation:
1Department of Creative Research, Exploratory Research Center on Life and Living Systems (ExCELLS), National Institutes of Natural Sciences, 5-1 Higashiyama, Myodaiji-cho, Okazaki 444-8787 Japan.