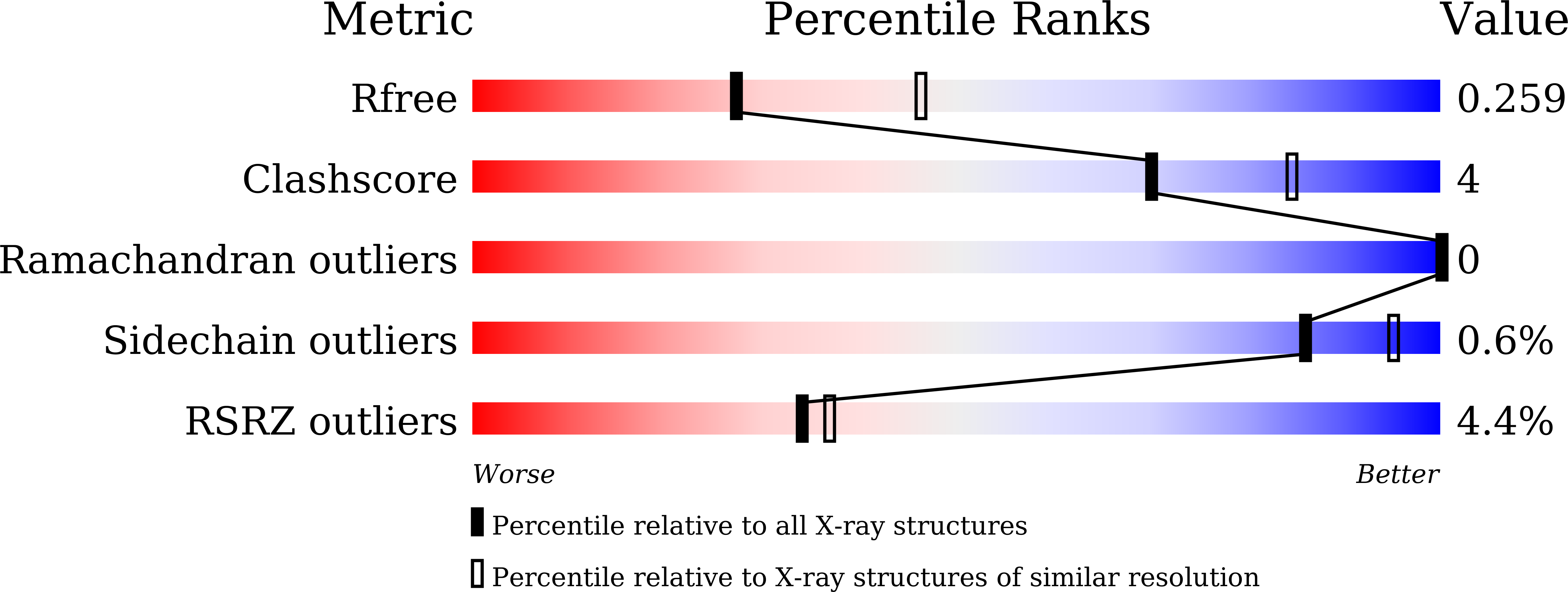
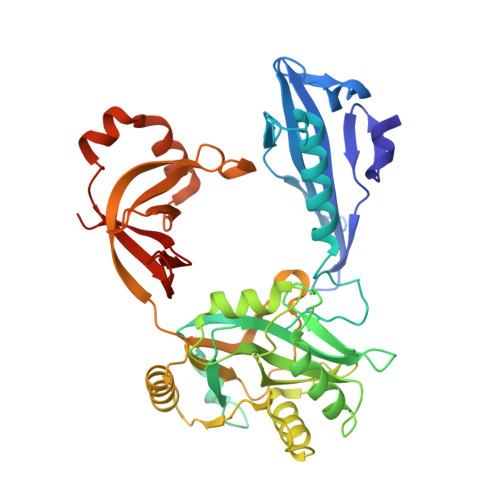
Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues.
Chen, Y., Liu, H., Yang, C., Gao, Y., Yu, X., Chen, X., Cui, R., Zheng, L., Li, S., Li, X., Ma, J., Huang, Z., Li, J., Gan, J.(2019) Nat Commun 10: 387-387
- PubMed: 30674878
- DOI: https://doi.org/10.1038/s41467-019-08296-w
- Primary Citation of Related Structures:
6IMJ, 6IMK, 6IML, 6IMN - PubMed Abstract:
African swine fever virus (ASFV) is contagious and can cause highly lethal disease in pigs. ASFV DNA ligase (AsfvLIG) is one of the most error-prone ligases identified to date; it catalyzes DNA joining reaction during DNA repair process of ASFV and plays important roles in mutagenesis of the viral genome. Here, we report four AsfvLIG:DNA complex structures and demonstrate that AsfvLIG has a unique N-terminal domain (NTD) that plays critical roles in substrate binding and catalytic complex assembly. In combination with mutagenesis, in vitro binding and catalytic assays, our study reveals that four unique active site residues (Asn153 and Leu211 of the AD domain; Leu402 and Gln403 of the OB domain) are crucial for the catalytic efficiency of AsfvLIG. These unique structural features can serve as potential targets for small molecule design, which could impair genome repair in ASFV and help combat this virus in the future.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Department of Physiology and Biophysics, School of Life Sciences, Fudan University, 200433, Shanghai, China.