Selection and analyses of variants of a designed protein suggest importance of hydrophobicity of partially buried sidechains for protein stability at high temperatures.
Han, M., Liao, S., Peng, X., Zhou, X., Chen, Q., Liu, H.(2019) Protein Sci 28: 1437-1447
- PubMed: 31074908
- DOI: https://doi.org/10.1002/pro.3643
- Primary Citation of Related Structures:
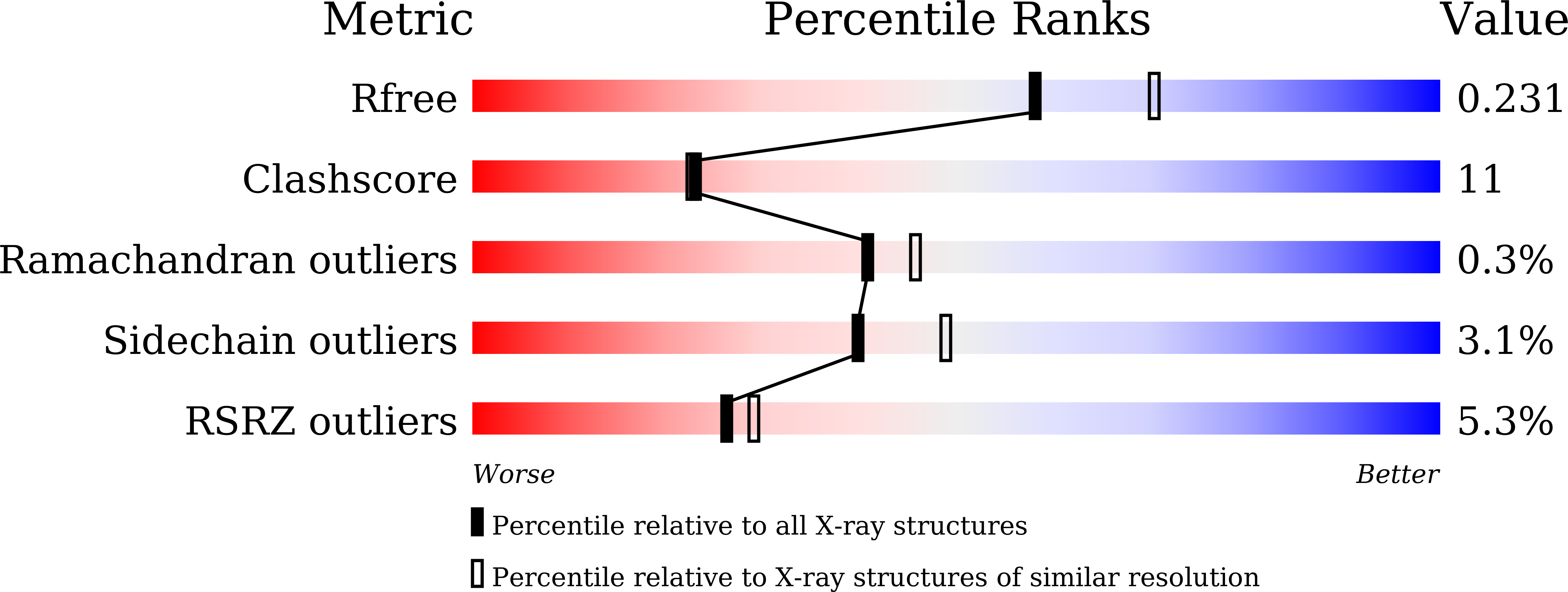
6IEV - PubMed Abstract:
Computationally designed proteins of high stability provide specimen in addition to natural proteins for the study of sequence-structure stability relationships at the very high end of protein stability spectrum. The melting temperature of E_1r26, a protein we previously designed using the A Backbone-based Amino aCid Usage Survey (ABACUS) sequence design program, is above 110 °C, more than 50 °C higher than that of the natural thioredoxin protein whose backbone (PDB ID 1R26) has been used as the design target. Using an experimental selection approach, we obtained variants of E_1r26 that remain folded but are of reduced stability, including one whose unfolding temperature and denaturing guanidine concentration are similar to those of 1r26. The mutant unfolds with a certain degree of cooperativity. Its structure solved by X-ray crystallography agrees with that of 1r26 by a root mean square deviation of 1.3 Å, adding supports to the accuracy of the ABACUS method. Analyses of intermediate mutants indicate that the substitution of two partially buried hydrophobic residues (isoleucine and leucine) by polar residues (threonine and serine, respectively) are responsible for the dramatic change in the unfolding temperature. It is suggested that the effects of mutations located in rigid secondary structure regions, but not those in loops, may be well predicted through ABACUS mutation energy analysis. The results also suggest that hydrophobic effects involving intermediately buried sidechains can be critically important for protein stability at high temperatures.
Organizational Affiliation:
School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, China.