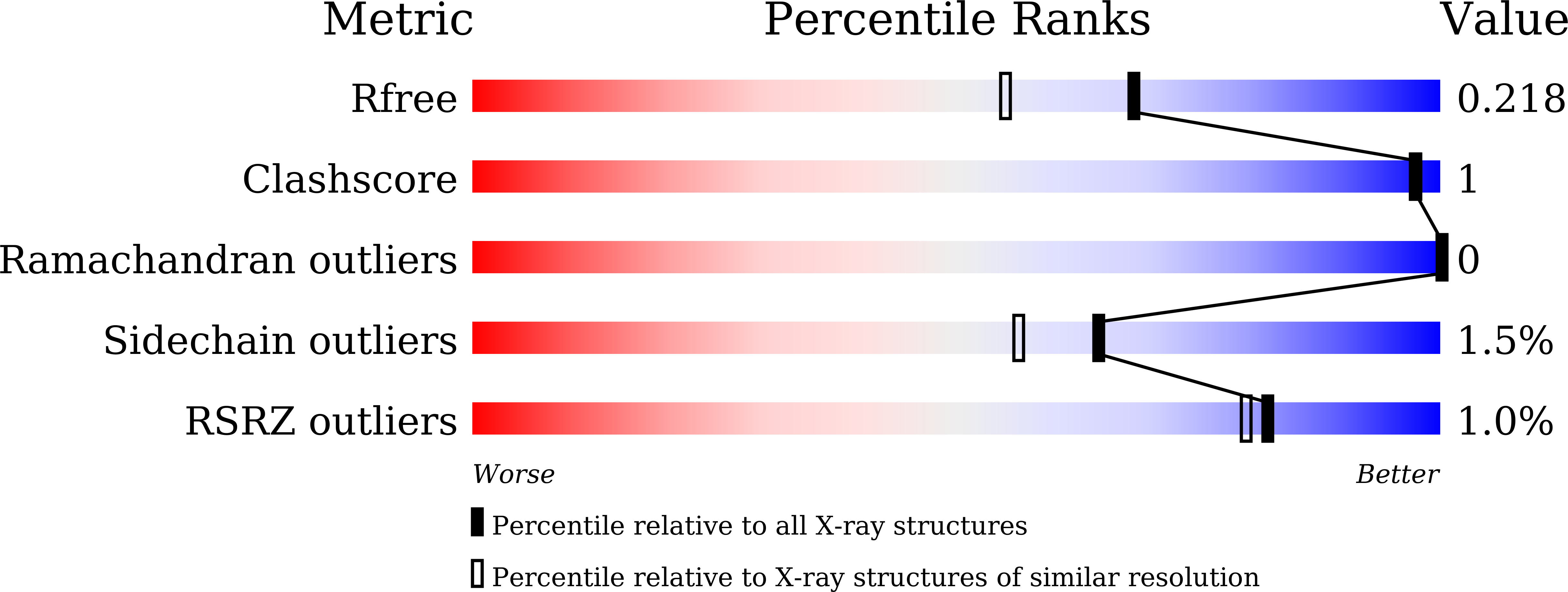
Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase.
Arakawa, T., Sato, Y., Yamada, M., Takabe, J., Yoshitaka Moriwaki, Y., Masamura, N., Kato, M., Aoyagi, M., Kamoi, T., Terada, T., Shimizu, K., Tsuge, N., Imai, S., Fushinobu, S.(2020) ACS Catal 10: 9-19