Structural, mechanistic, and physiological insights into phospholipase A-mediated membrane phospholipid degradation in Pseudomonas aeruginosa.
Bleffert, F., Granzin, J., Caliskan, M., Schott-Verdugo, S.N., Siebers, M., Thiele, B., Rahme, L., Felgner, S., Dormann, P., Gohlke, H., Batra-Safferling, R., Jaeger, K.E., Kovacic, F.(2022) Elife 11
- PubMed: 35536643
- DOI: https://doi.org/10.7554/eLife.72824
- Primary Citation of Related Structures:
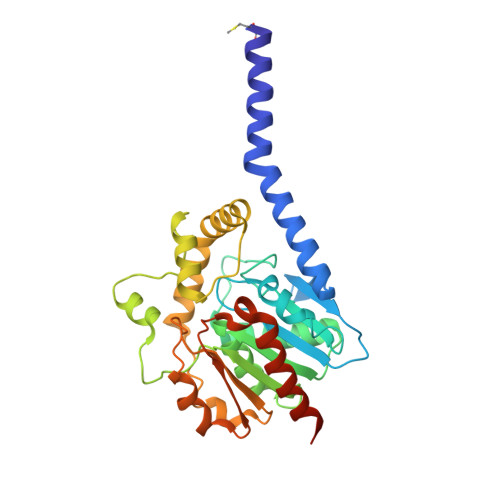
6I8W - PubMed Abstract:
Cells steadily adapt their membrane glycerophospholipid (GPL) composition to changing environmental and developmental conditions. While the regulation of membrane homeostasis via GPL synthesis in bacteria has been studied in detail, the mechanisms underlying the controlled degradation of endogenous GPLs remain unknown. Thus far, the function of intracellular phospholipases A (PLAs) in GPL remodeling (Lands cycle) in bacteria is not clearly established. Here, we identified the first cytoplasmic membrane-bound phospholipase A 1 (PlaF) from Pseudomonas aeruginosa , which might be involved in the Lands cycle. PlaF is an important virulence factor, as the P. aeruginosa Δ plaF mutant showed strongly attenuated virulence in Galleria mellonella and macrophages. We present a 2.0-Å-resolution crystal structure of PlaF, the first structure that reveals homodimerization of a single-pass transmembrane (TM) full-length protein. PlaF dimerization, mediated solely through the intermolecular interactions of TM and juxtamembrane regions, inhibits its activity. The dimerization site and the catalytic sites are linked by an intricate ligand-mediated interaction network, which might explain the product (fatty acid) feedback inhibition observed with the purified PlaF protein. We used molecular dynamics simulations and configurational free energy computations to suggest a model of PlaF activation through a coupled monomerization and tilting of the monomer in the membrane, which constrains the active site cavity into contact with the GPL substrates. Thus, these data show the importance of the PlaF-mediated GPL remodeling pathway for virulence and could pave the way for the development of novel therapeutics targeting PlaF.
Organizational Affiliation:
Institute of Molecular Enzyme Technology, Heinrich Heine University Düsseldorf, Forschungszentrum Jülich GmbH, Jülich, Germany.