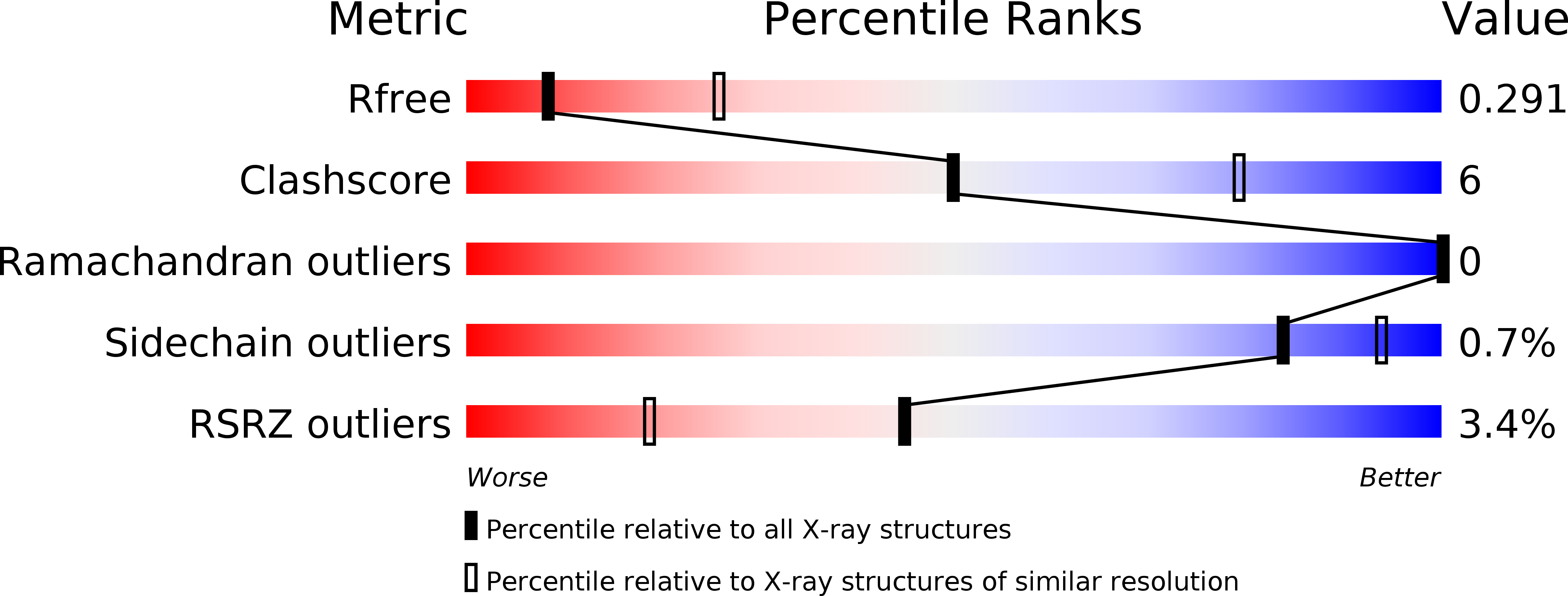
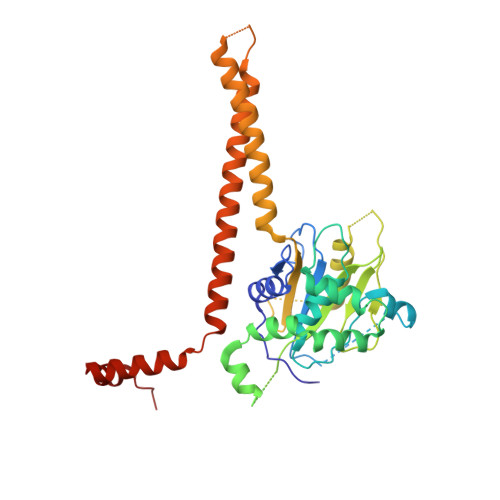
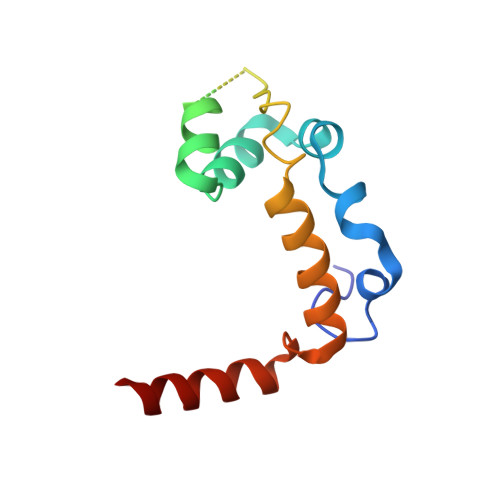
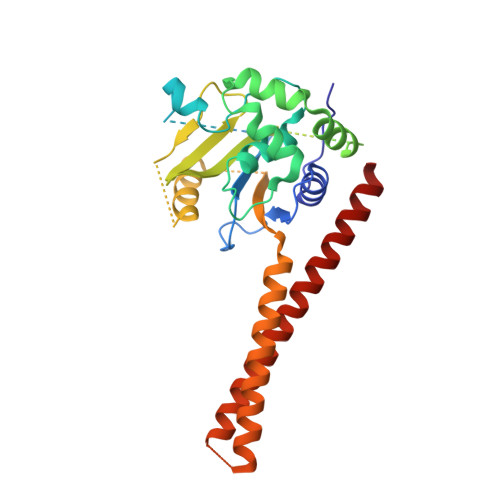
Structural Basis for the Activation of the Deubiquitinase Calypso by the Polycomb Protein ASX.
De, I., Chittock, E.C., Grotsch, H., Miller, T.C.R., McCarthy, A.A., Muller, C.W.(2019) Structure 27: 528-536.e4
- PubMed: 30639226
- DOI: https://doi.org/10.1016/j.str.2018.11.013
- Primary Citation of Related Structures:
6HGC - PubMed Abstract:
Ubiquitin C-terminal hydrolase deubiquitinase BAP1 is an essential tumor suppressor involved in cell growth control, DNA damage response, and transcriptional regulation. As part of the Polycomb repression machinery, BAP1 is activated by the deubiquitinase adaptor domain of ASXL1 mediating gene repression by cleaving ubiquitin (Ub) from histone H2A in nucleosomes. The molecular mechanism of BAP1 activation by ASXL1 remains elusive, as no structures are available for either BAP1 or ASXL1. Here, we present the crystal structure of the BAP1 ortholog from Drosophila melanogaster, named Calypso, bound to its activator, ASX, homolog of ASXL1. Based on comparative structural and functional analysis, we propose a model for Ub binding by Calypso/ASX, uncover decisive structural elements responsible for ASX-mediated Calypso activation, and characterize the interaction with ubiquitinated nucleosomes. Our results give molecular insight into Calypso function and its regulation by ASX and provide the opportunity for the rational design of mechanism-based therapeutics to treat human BAP1/ASXL1-related tumors.
Organizational Affiliation:
European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Meyerhofstrasse 1, 69117 Heidelberg, Germany.