The molybdenum storage protein - A bionanolab for creating experimentally alterable polyoxomolybdate clusters.
Brunle, S., Poppe, J., Hail, R., Demmer, U., Ermler, U.(2018) J Inorg Biochem 189: 172-179
Experimental Data Snapshot
(2018) J Inorg Biochem 189: 172-179
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Molybdenum storage protein subunit beta | A [auth B] | 269 | Azotobacter vinelandii | Mutation(s): 0 | 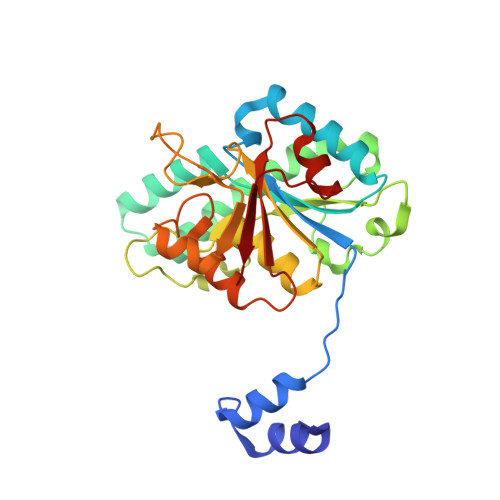 |
UniProt | |||||
Find proteins for P84253 (Azotobacter vinelandii (strain DJ / ATCC BAA-1303)) Explore P84253 Go to UniProtKB: P84253 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P84253 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Molybdenum storage protein subunit alpha | B [auth A] | 275 | Azotobacter vinelandii | Mutation(s): 0 | 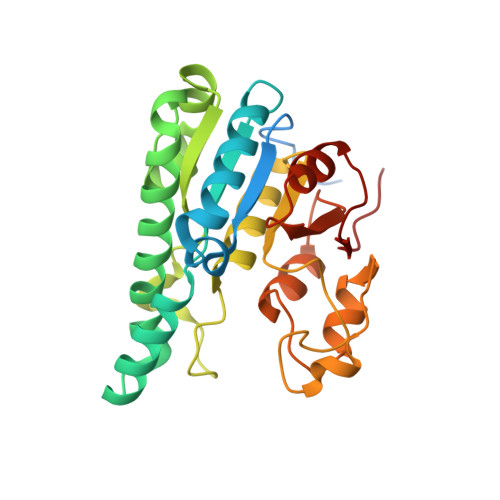 |
UniProt | |||||
Find proteins for P84308 (Azotobacter vinelandii (strain DJ / ATCC BAA-1303)) Explore P84308 Go to UniProtKB: P84308 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P84308 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 10 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| J7T Query on J7T | C [auth B] | molybdate cluster Mo9 O31 OPVBNKHERKYGEI-UHFFFAOYSA-A |  | ||
| J8B Query on J8B | J [auth A] | Molybdate cluster Mo8 O26 VDJFCWAFEZEUOL-UHFFFAOYSA-A |  | ||
| J7Q Query on J7Q | G [auth A] | tetrakis($l^{1}-oxidanyl)-[[2,2,2,4,4,4,4,6,6,6,8-undecakis($l^{1}-oxidanyl)-8-(oxidanylmolybdeniooxy)-6-[tris($l^{1}-oxidanyl)molybdeniooxy]-1,3,5,7-tetraoxa-2$l^{6},4$l^{6},6$l^{6},8$l^{4}-tetramolybdacyclooct-2-yl]oxy]molybdenum H Mo7 O26 JETHCXFIMFVFFB-UHFFFAOYSA-A |  | ||
| FUQ Query on FUQ | F [auth B] | Mo5 Cluster H20 Mo5 O25 KKTGFJKWMBVMER-UHFFFAOYSA-A |  | ||
| J85 Query on J85 | K [auth A] | [[[bis(oxidanylmolybdenio)-$l^{3}-oxidanyl]-$l^{1}-oxidanyl-oxidanylidene-molybdenio]-(oxidanylmolybdenio)-$l^{3}-oxidanyl]-tetrakis($l^{1}-oxidanyl)molybdenum H3 Mo5 O11 VWFFRZIWORKLLY-UHFFFAOYSA-F |  | ||
| J8E Query on J8E | E [auth B] | oxidanyl-[[2,2,4,4,4-pentakis($l^{1}-oxidanyl)-1-(oxidanylmolybdenio)-1$l^{3},3-dioxa-2$l^{5},4$l^{5}-dimolybdacyclobut-2-yl]oxy]molybdenum H2 Mo4 O10 DPQAEXIBFYBKAN-UHFFFAOYSA-G |  | ||
| ATP Query on ATP | H [auth A] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| J7N Query on J7N | D [auth B] | 2,2,4-tris(oxidanyl)-1,3-dioxa-2$l^{4},4$l^{3}-dimolybdacyclobutane H3 Mo2 O5 AUVZZTGQIKYYRK-UHFFFAOYSA-K |  | ||
| MOO Query on MOO | L [auth A] | MOLYBDATE ION Mo O4 MEFBJEMVZONFCJ-UHFFFAOYSA-N |  | ||
| MG Query on MG | I [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 116.72 | α = 90 |
| b = 116.72 | β = 90 |
| c = 235.42 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | phasing |