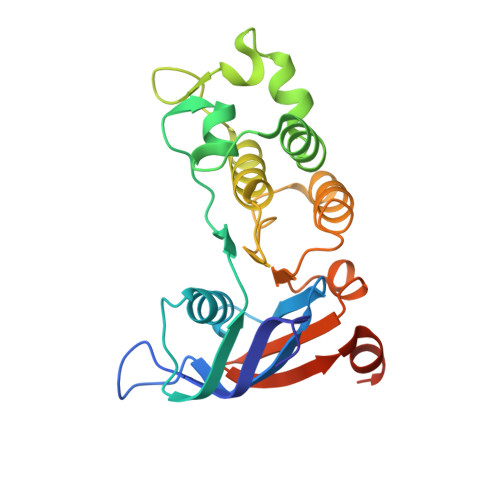
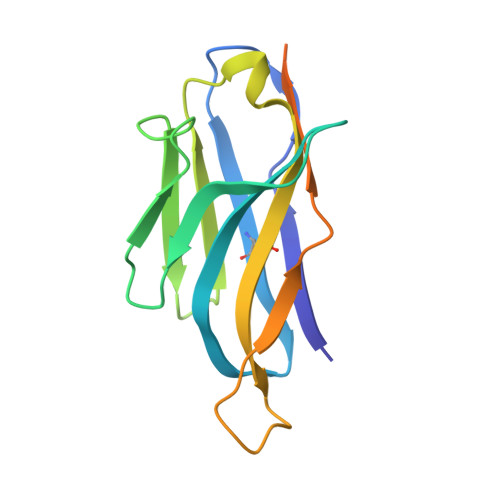
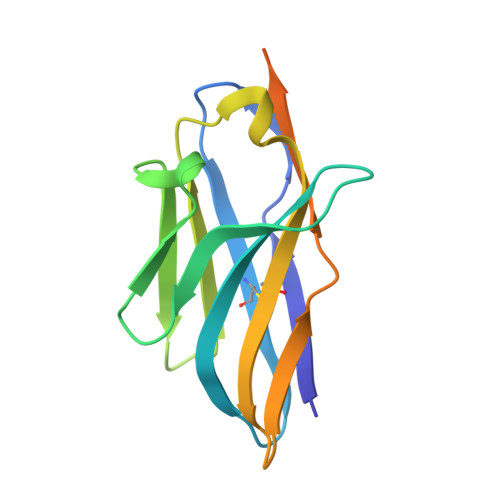
Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Sigoillot, M., Overtus, M., Grodecka, M., Scholl, D., Garcia-Pino, A., Laeremans, T., He, L., Pardon, E., Hildebrandt, E., Urbatsch, I., Steyaert, J., Riordan, J.R., Govaerts, C.(2019) Nat Commun 10: 2636-2636
- PubMed: 31201318
- DOI: https://doi.org/10.1038/s41467-019-10714-y
- Primary Citation of Related Structures:
6GJQ, 6GJS, 6GJU, 6GK4, 6GKD - PubMed Abstract:
The leading cause of cystic fibrosis (CF) is the deletion of phenylalanine 508 (F508del) in the first nucleotide-binding domain (NBD1) of the cystic fibrosis transmembrane conductance regulator (CFTR). The mutation affects the thermodynamic stability of the domain and the integrity of the interface between NBD1 and the transmembrane domain leading to its clearance by the quality control system. Here, we develop nanobodies targeting NBD1 of human CFTR and demonstrate their ability to stabilize both isolated NBD1 and full-length protein. Crystal structures of NBD1-nanobody complexes provide an atomic description of the epitopes and reveal the molecular basis for stabilization. Furthermore, our data uncover a conformation of CFTR, involving detachment of NBD1 from the transmembrane domain, which contrast with the compact assembly observed in cryo-EM structures. This unexpected interface rearrangement is likely to have major relevance for CF pathogenesis but also for the normal function of CFTR and other ABC proteins.
Organizational Affiliation:
SFMB, Université Libre de Bruxelles (ULB), CP206/02, Boulevard du Triomphe, building BC, B-1050, Brussels, Belgium.