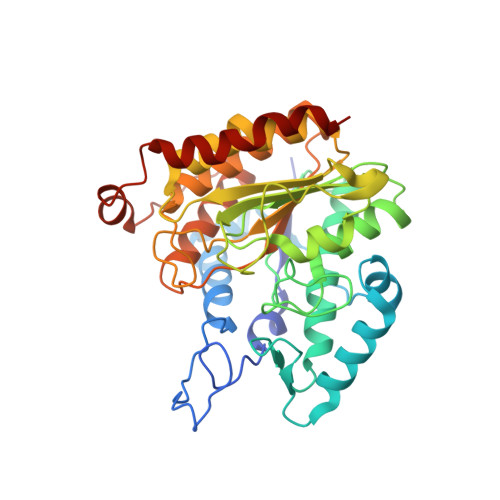
Covalent inhibition of histone deacetylase 8 by 3,4-dihydro-2H-pyrimido[1,2-c][1,3]benzothiazin-6-imine.
Muth, M., Jansch, N., Kopranovic, A., Kramer, A., Wossner, N., Jung, M., Kirschhofer, F., Meyer-Almes, F.J.(2019) Biochim Biophys Acta Gen Subj 1863: 577-585
- PubMed: 30611847
- DOI: https://doi.org/10.1016/j.bbagen.2019.01.001
- Primary Citation of Related Structures:
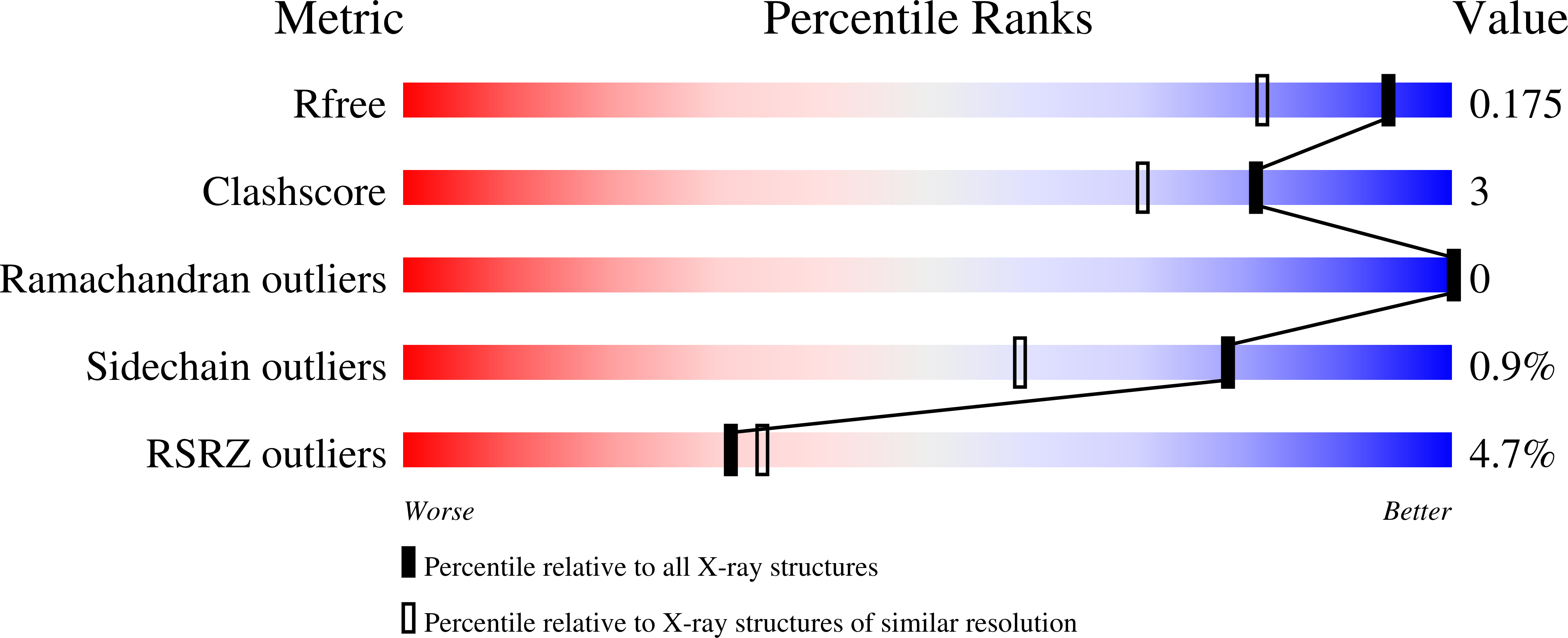
6GJK - PubMed Abstract:
HDAC8 is an established target for T-cell lymphoma and childhood neuroblastoma. Benzothiazine-imines are promising HDAC8 inhibitors with unknown binding mechanism lacking a usual zinc binding group.
Organizational Affiliation:
Department of Chemical Engineering and Biotechnology, University of Applied Sciences Darmstadt, Germany; Bioengineering and Biosystems, Institute of Functional Interfaces, Karlsruhe Institute of Technology, Germany.