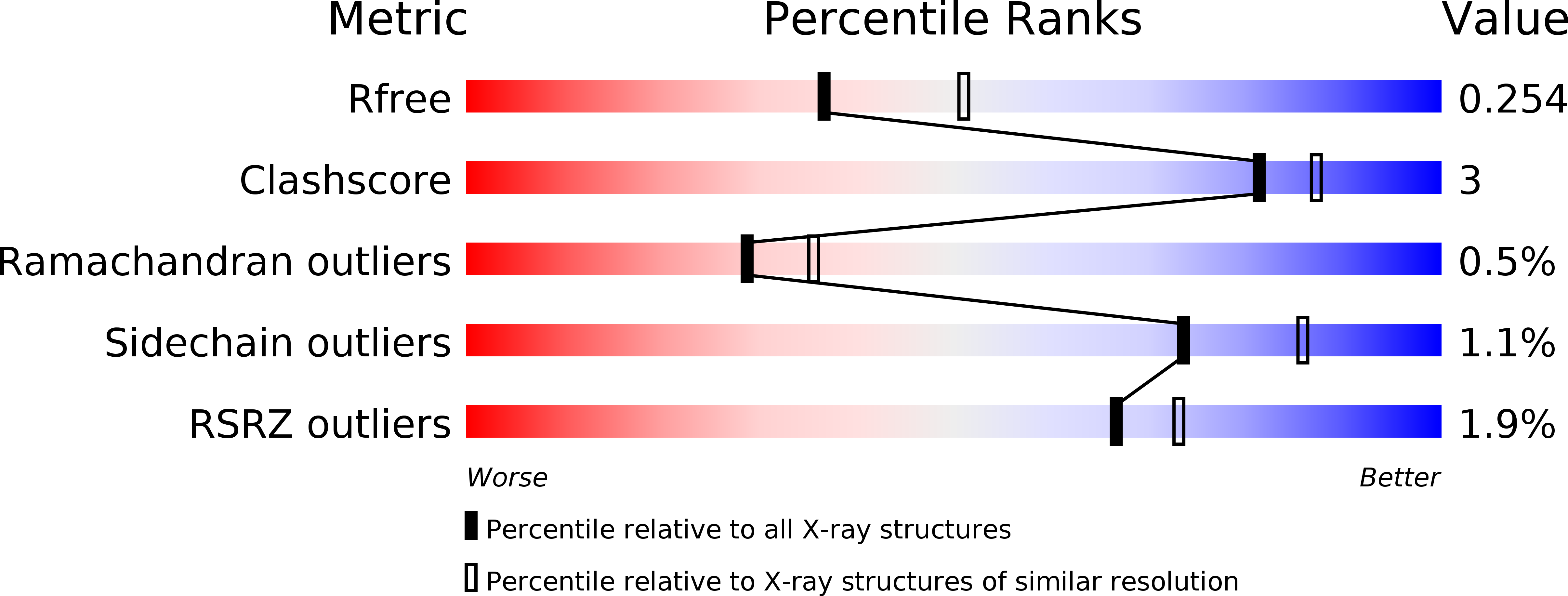
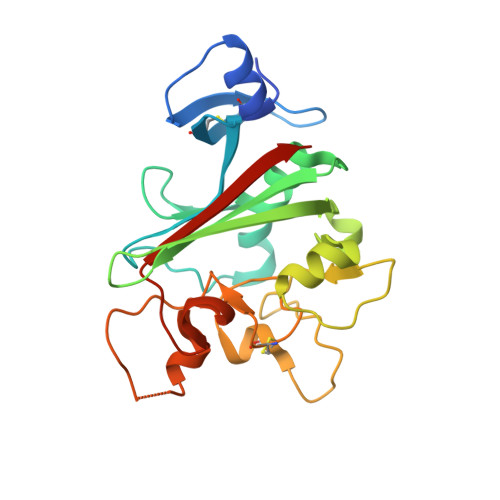
Structures of Angptl3 and Angptl4, modulators of triglyceride levels and coronary artery disease.
Biterova, E., Esmaeeli, M., Alanen, H.I., Saaranen, M., Ruddock, L.W.(2018) Sci Rep 8: 6752-6752
- PubMed: 29713054
- DOI: https://doi.org/10.1038/s41598-018-25237-7
- Primary Citation of Related Structures:
6EUA, 6EUB - PubMed Abstract:
Coronary artery disease is the most common cause of death globally and is linked to a number of risk factors including serum low density lipoprotein, high density lipoprotein, triglycerides and lipoprotein(a). Recently two proteins, angiopoietin-like protein 3 and 4, have emerged from genetic studies as being factors that significantly modulate plasma triglyceride levels and coronary artery disease. The exact function and mechanism of action of both proteins remains to be elucidated, however, mutations in these proteins results in up to 34% reduction in coronary artery disease and inhibition of function results in reduced plasma triglyceride levels. Here we report the crystal structures of the fibrinogen-like domains of both proteins. These structures offer new insights into the reported loss of function mutations, the mechanisms of action of the proteins and open up the possibility for the rational design of low molecular weight inhibitors for intervention in coronary artery disease.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Biology and Biocenter Oulu, University of Oulu, Oulu, 90220, Finland.