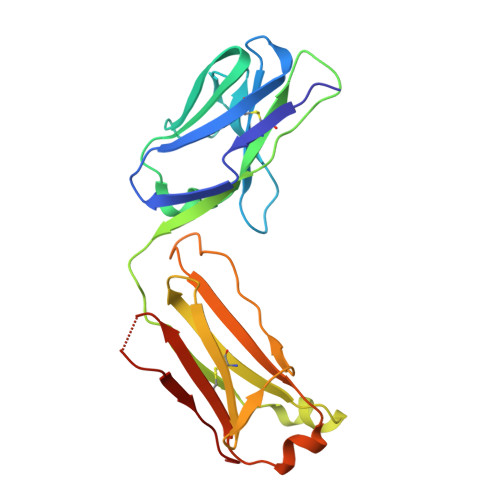
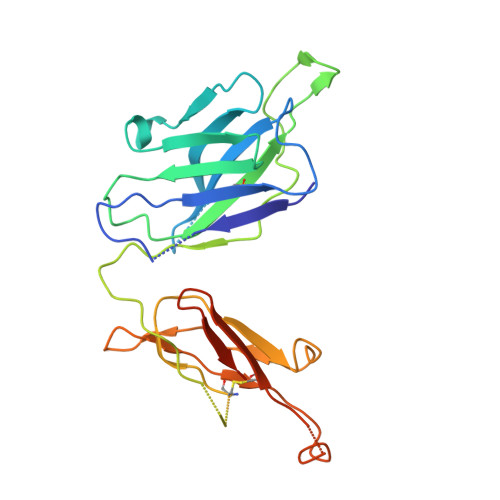
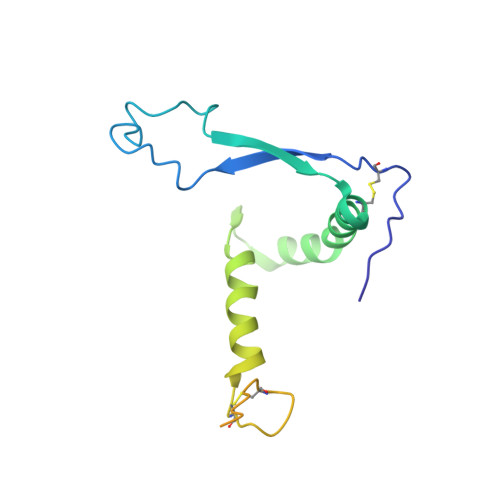
Structural basis for broad neutralization of ebolaviruses by an antibody targeting the glycoprotein fusion loop.
Janus, B.M., van Dyk, N., Zhao, X., A Howell, K., Soto, C., Aman, M.J., Li, Y., Fuerst, T.R., Ofek, G.(2018) Nat Commun 9: 3934-3934
- PubMed: 30258051
- DOI: https://doi.org/10.1038/s41467-018-06113-4
- Primary Citation of Related Structures:
6EAY - PubMed Abstract:
The severity of the 2014-2016 ebolavirus outbreak in West Africa expedited clinical development of therapeutics and vaccines though the countermeasures on hand were largely monospecific and lacked efficacy against other ebolavirus species that previously emerged. Recent studies indicate that ebolavirus glycoprotein (GP) fusion loops are targets for cross-protective antibodies. Here we report the 3.72 Å resolution crystal structure of one such cross-protective antibody, CA45, bound to the ectodomain of Ebola virus (EBOV) GP. The CA45 epitope spans multiple faces of the fusion loop stem, across both GP1 and GP2 subunits, with ~68% of residues identical across > 99.5% of known ebolavirus isolates. Extensive antibody interactions within a pan-ebolavirus small-molecule inhibitor binding cavity on GP define this cavity as a novel site of immune vulnerability. The structure elucidates broad ebolavirus neutralization through a highly conserved epitope on GP and further enables rational design and development of broadly protective vaccines and therapeutics.
Organizational Affiliation:
Institute for Bioscience and Biotechnology Research, University of Maryland, Rockville, MD, 20850, USA.