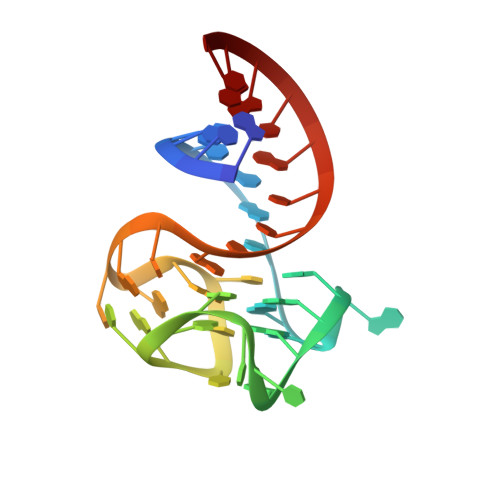
Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Trachman 3rd., R.J., Autour, A., Jeng, S.C.Y., Abdolahzadeh, A., Andreoni, A., Cojocaru, R., Garipov, R., Dolgosheina, E.V., Knutson, J.R., Ryckelynck, M., Unrau, P.J., Ferre-D'Amare, A.R.(2019) Nat Chem Biol 15: 472-479
- PubMed: 30992561
- DOI: https://doi.org/10.1038/s41589-019-0267-9
- Primary Citation of Related Structures:
6E8S, 6E8T, 6E8U - PubMed Abstract:
Several turn-on RNA aptamers that activate small-molecule fluorophores have been selected in vitro. Among these, the ~30 nucleotide Mango-III is notable because it binds the thiazole orange derivative TO1-Biotin with high affinity and fluoresces brightly (quantum yield 0.55). Uniquely among related aptamers, Mango-III exhibits biphasic thermal melting, characteristic of molecules with tertiary structure. We report crystal structures of TO1-Biotin complexes of Mango-III, a structure-guided mutant Mango-III(A10U), and a functionally reselected mutant iMango-III. The structures reveal a globular architecture arising from an unprecedented pseudoknot-like connectivity between a G-quadruplex and an embedded non-canonical duplex. The fluorophore is restrained into a planar conformation by the G-quadruplex, a lone, long-range trans Watson-Crick pair (whose A10U mutation increases quantum yield to 0.66), and a pyrimidine perpendicular to the nucleobase planes of those motifs. The improved iMango-III and Mango-III(A10U) fluoresce ~50% brighter than enhanced green fluorescent protein, making them suitable tags for live cell RNA visualization.
Organizational Affiliation:
Biochemistry and Biophysics Center, National Heart, Lung, and Blood Institute, Bethesda, MD, USA.