Crystal structure of LpxC from Pseudomonas aeruginosa in complex with ligand PT803
Delker, S.L., Mayclin, S.J., Phan, J.N., Abendroth, J., Lorimer, D., Edwards, T.E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase | 304 | Pseudomonas aeruginosa UCBPP-PA14 | Mutation(s): 1 Gene Names: lpxC, PA14_57260 EC: 3.5.1.108 | 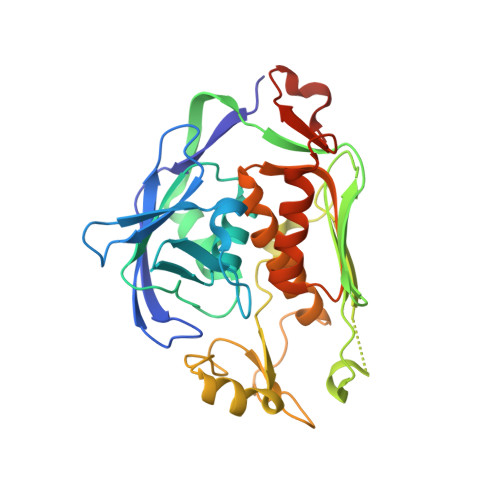 | |
UniProt | |||||
Find proteins for Q02H34 (Pseudomonas aeruginosa (strain UCBPP-PA14)) Explore Q02H34 Go to UniProtKB: Q02H34 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02H34 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| F64 Query on F64 | C [auth A] | (2S)-4-[4-{4-[(5-chloro-6-methoxypyridin-3-yl)methoxy]phenyl}-2-oxo-3,6-dihydropyridin-1(2H)-yl]-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide C24 H28 Cl N3 O7 S CDFIGPHKVBKXMP-DEOSSOPVSA-N |  | ||
| EU1 Query on EU1 | B [auth A] | (2R)-4-[4-{4-[(5-chloro-6-methoxypyridin-3-yl)methoxy]phenyl}-2-oxo-3,6-dihydropyridin-1(2H)-yl]-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide C24 H28 Cl N3 O7 S CDFIGPHKVBKXMP-XMMPIXPASA-N |  | ||
| ZN Query on ZN | D [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | E [auth A], F [auth A], G [auth A], H [auth A], I [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 35.66 | α = 90 |
| b = 79.34 | β = 92.01 |
| c = 50.92 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XSCALE | data scaling |
| PDB_EXTRACT | data extraction |
| MOLREP | phasing |