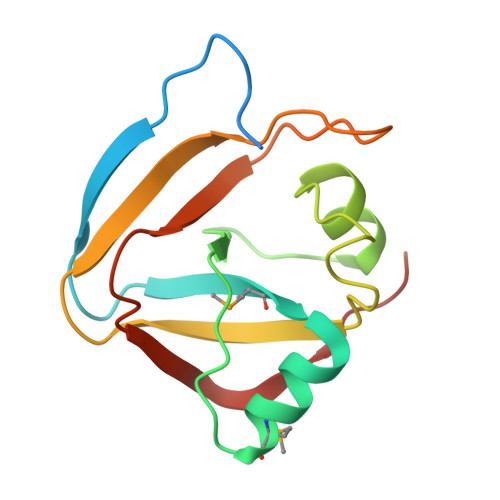
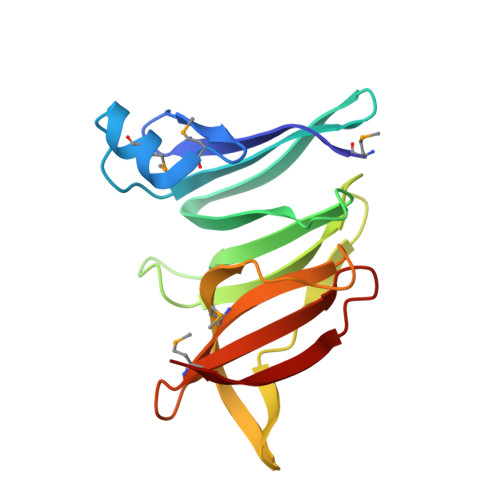
Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system.
Tang, J.Y., Bullen, N.P., Ahmad, S., Whitney, J.C.(2018) J Biol Chem 293: 1504-1514
- PubMed: 29237732
- DOI: https://doi.org/10.1074/jbc.RA117.000178
- Primary Citation of Related Structures:
6B12 - PubMed Abstract:
The bacterial type VI secretion system (T6SS) mediates antagonistic cell-cell interactions between competing Gram-negative bacteria. In plant-beneficial bacteria, this pathway has been shown to suppress the growth of bacterial pathogens; however, the identification and mode of action of T6SS effector proteins that mediate this protective effect remain poorly defined. Here, we identify two previously uncharacterized effectors required for interbacterial antagonism by the plant commensal bacterium Pseudomonas protegens Consistent with the established effector-immunity paradigm for antibacterial T6SS substrates, the toxic activities of these effectors are neutralized by adjacently encoded cognate immunity determinants. Although one of these effectors, RhsA, belongs to the family of DNase enzymes, the activity of the other was not apparent from its sequence. To determine the mechanism of toxicity of this latter effector, we determined its 1.3 Å crystal structure in complex with its immunity protein and found that it resembles NAD(P) + -degrading enzymes. In line with this structural similarity, biochemical characterization of this effector, termed Tne2 ( T ype VI secretion N ADase e ffector family 2), demonstrates that it possesses potent NAD(P) + hydrolase activity. Tne2 is the founding member of a widespread family of interbacterial NADases predicted to transit not only the Gram-negative T6SS but also the Gram-positive type VII secretion system, a pathway recently implicated in interbacterial competition among Firmicutes. Together, this work identifies new T6SS effectors employed by a plant commensal bacterium to antagonize its competitors and broadly implicates NAD(P) + -hydrolyzing enzymes as substrates of interbacterial conflict pathways found in diverse bacterial phyla.
Organizational Affiliation:
From the Michael DeGroote Institute for Infectious Disease Research and.