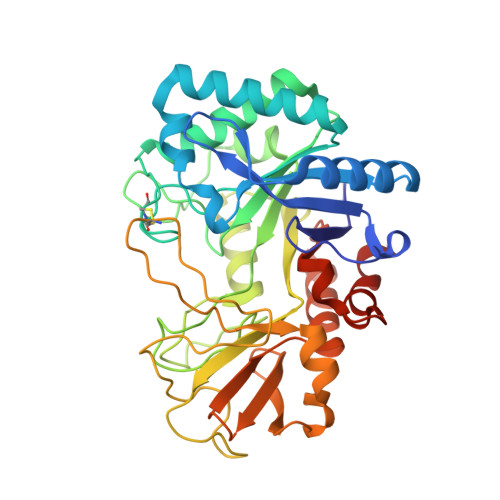
Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Jiang, Z., Hu, S., Ma, J., Liu, Y., Qiao, Z., Yan, Q., Gao, Y., Yang, S.(2021) Biochim Biophys Acta Proteins Proteom : 140709-140709
- PubMed: 34358705
- DOI: https://doi.org/10.1016/j.bbapap.2021.140709
- Primary Citation of Related Structures:
5XWF, 5YUQ, 7FBT - PubMed Abstract:
A chitinase gene (RmChiA) encoding 445 amino acid (aa) residues from a fungus Rhizomucor miehei was cloned and overexpressed in Escherichia coli. Two kinds of RmChiA crystal forms, with space groups P3 2 2 1 and P1, were obtained by sitting-drop vapor diffusion and the structures were determined by X-ray diffraction. The overall structure of RmChiA monomer, which is the first structure of bacterial-type chitinases from nonpathogenic fungi, adopts a canonical triosephosphate isomerase (TIM) barrel fold with two protruding chitinase insertion domains. RmChiA exhibited a unique NxDxE catalytical motif and a real active site tunnel structure, which are firstly found in GH family 18 chitinases. The motif had high structural homolog with the typical DxDxE motif in other GH family 18 chitinases. The tunnel is formed by two unusual long loops, containing 15 aa and 45 aa respectively, linked by a disulfide bond across the substrate-binding cleft. Mutation experiments found that opening the roof of tunnel structure increased the hydrolysis efficiency of RmChiA, but the thermostability of the mutants decreased. Moreover, the tunnel structure endowed RmChiA with the exo-chitinase character.
Organizational Affiliation:
College of Food Science and Nutritional Engineering, China Agricultural University, Beijing 100083, China.