Structural and Biochemical Study of the Mono-ADP-Ribosyltransferase Domain of SdeA, a Ubiquitylating/Deubiquitylating Enzyme from Legionella pneumophila
Kim, L., Kwon, D.H., Kim, B.H., Kim, J., Park, M.R., Park, Z.Y., Song, H.K.(2018) J Mol Biol 430: 2843-2856
- PubMed: 29870726
- DOI: https://doi.org/10.1016/j.jmb.2018.05.043
- Primary Citation of Related Structures:
5YSI, 5YSJ, 5YSK - PubMed Abstract:
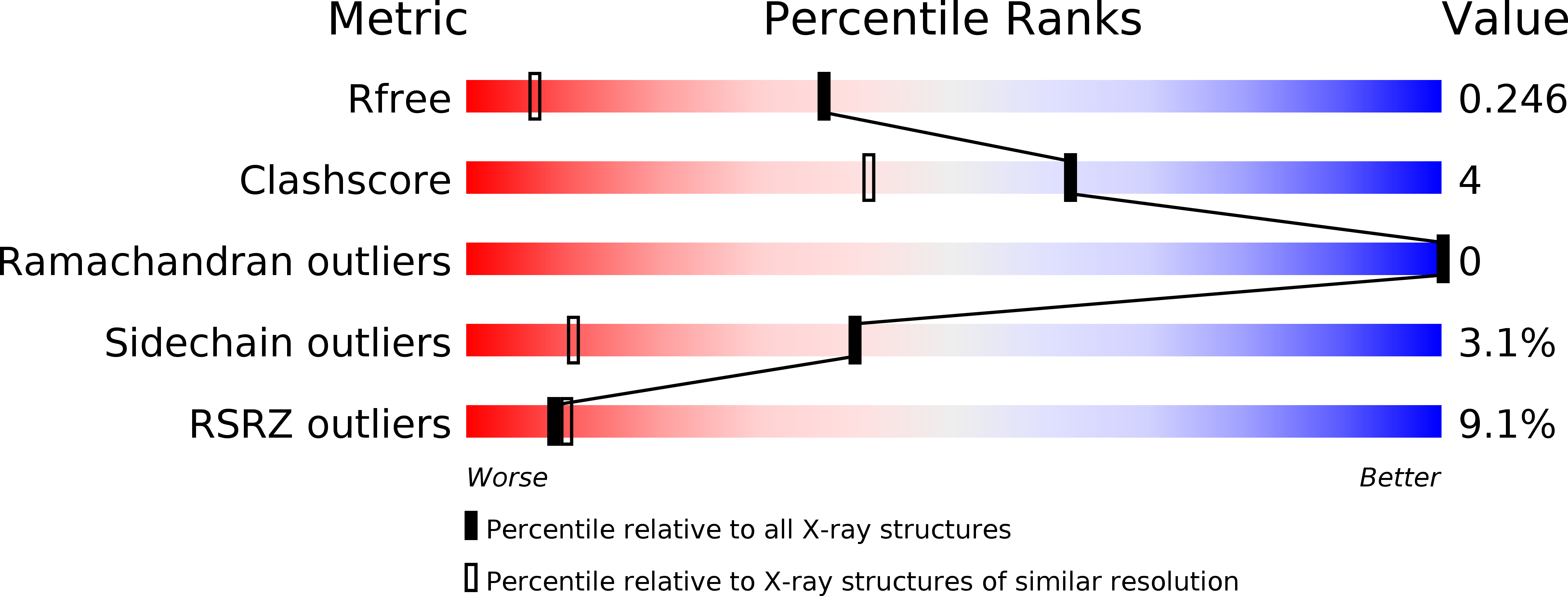
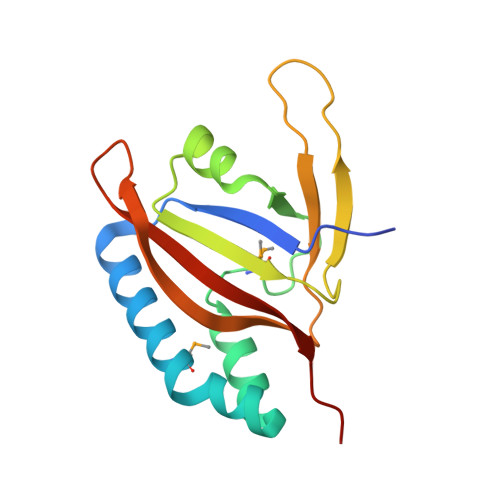
Conventional ubiquitylation occurs through an ATP-dependent three-enzyme cascade (E1, E2, and E3) that mediates the covalent conjugation of the C-terminus of ubiquitin to a lysine on the substrate. SdeA, which belongs to the SidE effector family of Legionella pneumophila, can transfer ubiquitin to endoplasmic reticulum-associated Rab-family GTPases in a manner independent of E1 and E2 enzymes. The novel ubiquitin-modifying enzyme SdeA utilizes NAD + as a cofactor to attach ubiquitin to a serine residue of the substrate. Here, to elucidate the coupled enzymatic reaction of NAD+ hydrolysis and ADP-ribosylation of ubiquitin in SdeA, we characterized the mono-ADP-ribosyltransferase domain of SdeA and show that it consists of two sub-domains termed mART-N and mART-C. The crystal structure of the mART-C domain of SdeA was also determined in free form and in complex with NAD + at high resolution. Furthermore, the spatial orientations of the N-terminal deubiquitylase, phosphodiesterase, mono-ADP-ribosyltransferase, and C-terminal coiled-coil domains within the 180-kDa full-length SdeA were determined. These results provide insight into the unusual ubiquitylation mechanism of SdeA and expand our knowledge on the structure-function of mono-ADP-ribosyltransferases.
Organizational Affiliation:
Department of Life Sciences, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, Korea.