Crystal structure of the post-fusion core of the Human coronavirus 229E spike protein at 1.86 angstrom resolution
Yan, L., Meng, B., Xiang, J., Wilson, I.A., Yang, B.(2018) Acta Crystallogr D Struct Biol 74: 841-851
- PubMed: 30198895
- DOI: https://doi.org/10.1107/S2059798318008318
- Primary Citation of Related Structures:
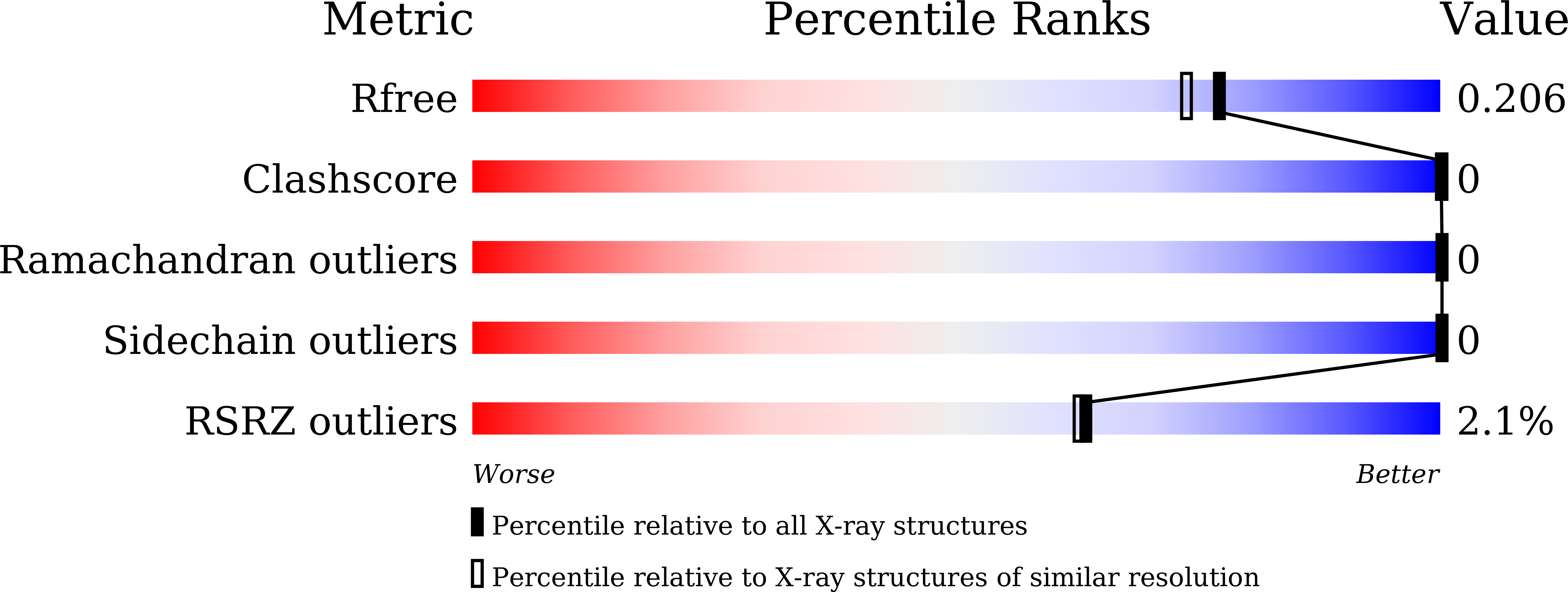
5YL9 - PubMed Abstract:
Human coronavirus 229E (HCoV-229E) usually causes mild upper respiratory infections in heathy adults, but may lead to severe complications or mortality in individuals with weakened immune systems. Virus entry of HCoV-229E is mediated by its spike (S) protein, where the S1 domain facilitates attachment to host cells and the S2 domain is involved in subsequent fusion of the virus and host membranes. During the fusion process, two heptad repeats, HR1 and HR2, in the S2 domain assemble into a six-helix membrane-fusion structure termed the fusion core. Here, the complete fusion-core structure of HCoV-229E has been determined at 1.86 Å resolution, representing the most complete post-fusion conformation thus far among published human alphacoronavirus (α-HCoV) fusion-core structures. The overall structure of the HCoV-229E fusion core is similar to those of SARS, MERS and HCoV-NL63, but the packing of its 3HR1 core differs from those of SARS and MERS in that it contains more noncanonical `x' and `da' layers. Side-by-side electrostatic surface comparisons reveal that the electrostatic surface potentials are opposite in α-HCoVs and β-HCoVs at certain positions and that the HCoV-229E surface also appears to be the most hydrophobic among the various HCoVs. In addition to the highly conserved hydrophobic interactions between HR1 and HR2, some polar and electrostatic interactions are also well preserved across different HCoVs. This study adds to the structural profiling of HCoVs to aid in the structure-based design of pan-coronavirus small molecules or peptides to inhibit viral fusion.
Organizational Affiliation:
Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, 393 Middle Huaxia Road, Shanghai 201210, People's Republic of China.