Structural and binding studies of phosphopantetheine adenylyl transferase from Acinetobacter baumannii.
Gupta, A., Singh, P.K., Iqbal, N., Sharma, P., Baraigya, H.R., Kaur, P., Umar, M.S., Ahmad, F., Sharma, A., Owais, M., Sharma, S., Singh, T.P.(2019) Biochim Biophys Acta Proteins Proteom 1867: 537-547
- PubMed: 30885618
- DOI: https://doi.org/10.1016/j.bbapap.2019.03.002
- Primary Citation of Related Structures:
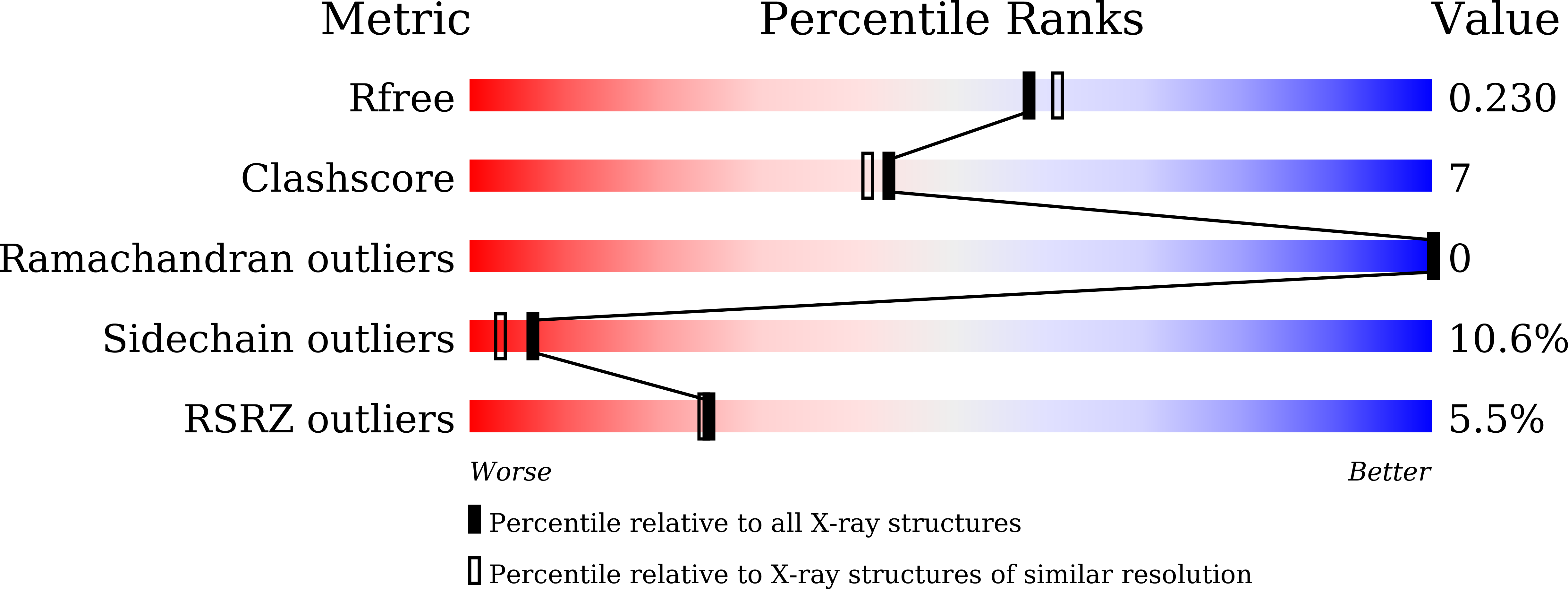
5H7X, 5YH7 - PubMed Abstract:
Phosphopantetheine adenylyltransferase (PPAT, EC. 2.7.7.3) catalyzes an essential step in the reaction that transfers an adenylyl group from adenosine tri phosphate (ATP) to 4'-phosphopantetheine (pPant) yielding 3'- dephospho-coenzyme A (dPCoA) and pyrophosphate (PP) in the coenzyme A (CoA) biosynthesis pathway . The enzyme PPAT from Acinetobacter baumannii (AbPPAT) was cloned, expressed and purified. The binding studies of AbPPAT were carried out with two compounds, tri‑sodium citrate (TSC) and l-ascorbic acid (LAA, vitamin-C) using fluorescence spectroscopic (FS) and surface Plasmon resonance (SPR) methods. Both methods provided similar values of dissociation constants for TSC and LAA which were of the order of 10 -8 M and 10 -5 M respectively. The computer aided docking studies indicated fewer interactions of LAA with AbPPAT as compared to those of TSC. The freshly purified samples of AbPPAT were crystallized. The crystals of AbPPAT were soaked in the solutions containing TSC and LAA. However, the crystals of the complex of AbPPAT with LAA did not diffract well and hence the structure of the complex of AbPPAT with LAA could not be determined. On the other hand, the crystals of the complex of AbPPAT with TSC diffracted well and the structure was determined at 1.76 Å resolution. It showed that TSC bound to AbPPAT at the ATP binding site and formed several intermolecular contacts including 12 hydrogen bonds. The results of binding studies for both TSC and LAA and the structure of the complex of AbPPAT with TSC clearly indicated a potential role of TSC and LAA as antibacterial agents.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.