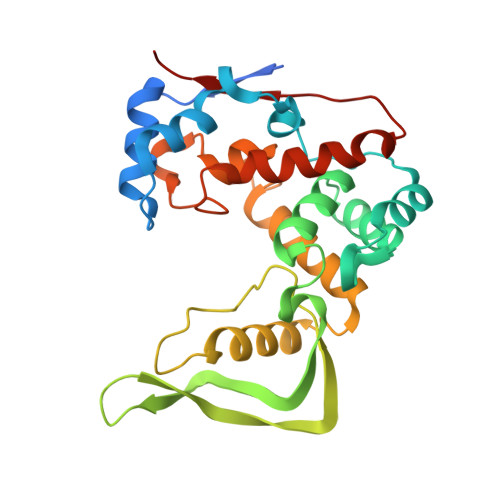
Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain
Tamai, E., Sekiya, H., Goda, E., Makihata, N., Maki, J., Yoshida, H., Kamitori, S.(2017) FEBS Lett 591: 231-239
- PubMed: 27926788
- DOI: https://doi.org/10.1002/1873-3468.12515
- Primary Citation of Related Structures:
5WQW - PubMed Abstract:
Bacterial autolysins can partially hydrolyze cell wall peptidoglycans into small sections to regulate cell separation/division and the growth phase. Clostridium perfringens autolysin (Acp) has an N-terminal cell wall-binding domain and a C-terminal catalytic domain with glucosaminidase activity that belongs to the glycoside hydrolase 73 family. Here, we determined the X-ray structure of the Acp catalytic domain (AcpCD) at 1.76 Å resolution. AcpCD has a unique crescent-shaped structure, forming a deep groove for substrate-binding at the center of the protein. The modeling study of the enzyme/substrate complex demonstrated that the length of the substrate-binding groove is closely related to the glucosaminidase activity. Mutagenesis analysis showed that AcpCD likely adopts a neighboring-group mechanism for the catalytic reaction.
Organizational Affiliation:
Department of Infectious Disease, College of Pharmaceutical Science, Matsuyama University, Bunkyo-cho, Ehime, Japan.