Reducing Macro- and Microheterogeneity of N-Glycans Enables the Crystal Structure of the Lectin and EGF-Like Domains of Human L-Selectin To Be Solved at 1.9 angstrom Resolution.
Wedepohl, S., Dernedde, J., Vahedi-Faridi, A., Tauber, R., Saenger, W., Bulut, H.(2017) Chembiochem 18: 1338-1345
- PubMed: 28489325
- DOI: https://doi.org/10.1002/cbic.201700220
- Primary Citation of Related Structures:
5VC1 - PubMed Abstract:
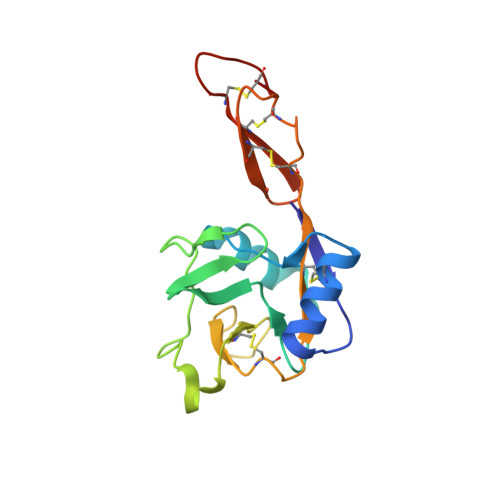
L-Selectin, a cell-adhesion receptor on the surface of most leukocytes, contains seven N-glycosylation sites. In order to obtain the crystal structure of human L-selectin, we expressed a shortened version of L-selectin comprising the C-type lectin and EGF-like domains (termed LE) and systematically analysed mutations of the three glycosylation sites (Asn22, Asn66 and Asn139) in order to reduce macroheterogeneity. After we further removed microheterogeneity, we obtained crystals that diffracted X-rays up to 1.9 Å from a variant (LE010) with exchanges N22Q and N139Q and one GlcNAc 2 Man 5 N-glycan chain attached to Asn66. Crystal-structure analysis showed that the terminal mannose of GlcNAc 2 Man 5 of one LE010 molecule was coordinated to Ca 2+ in the binding site of a symmetry-related LE010. The orientation of the lectin and EGF-like domain was similar to the described "bent" conformation of E- and P-selectins. The Ca 2+ -binding site reflects the binding mode seen in E- and P-selectin structures co-crystallised with ligands.
Organizational Affiliation:
Institut für Laboratoriumsmedizin, Klinische Chemie und Pathobiochemie, Charité Universitätsmedizin Berlin, CVK, Augustenburger Platz 1, 13353, Berlin, Germany.