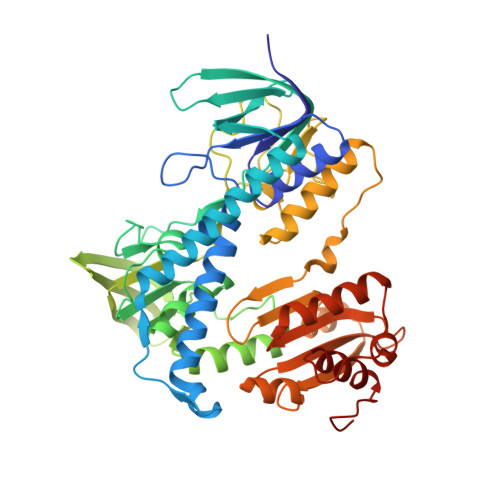
2.5 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD.
Minasov, G., Wawrzak, Z., Shuvalova, L., Kiryukhina, O., Dubrovska, I., Grimshaw, S., Kwon, K., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.