7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
Bruender, N.A., Grell, T.A., Dowling, D.P., McCarty, R.M., Drennan, C.L., Bandarian, V.(2017) J Am Chem Soc 139: 1912-1920
- PubMed: 28045519
- DOI: https://doi.org/10.1021/jacs.6b11381
- Primary Citation of Related Structures:
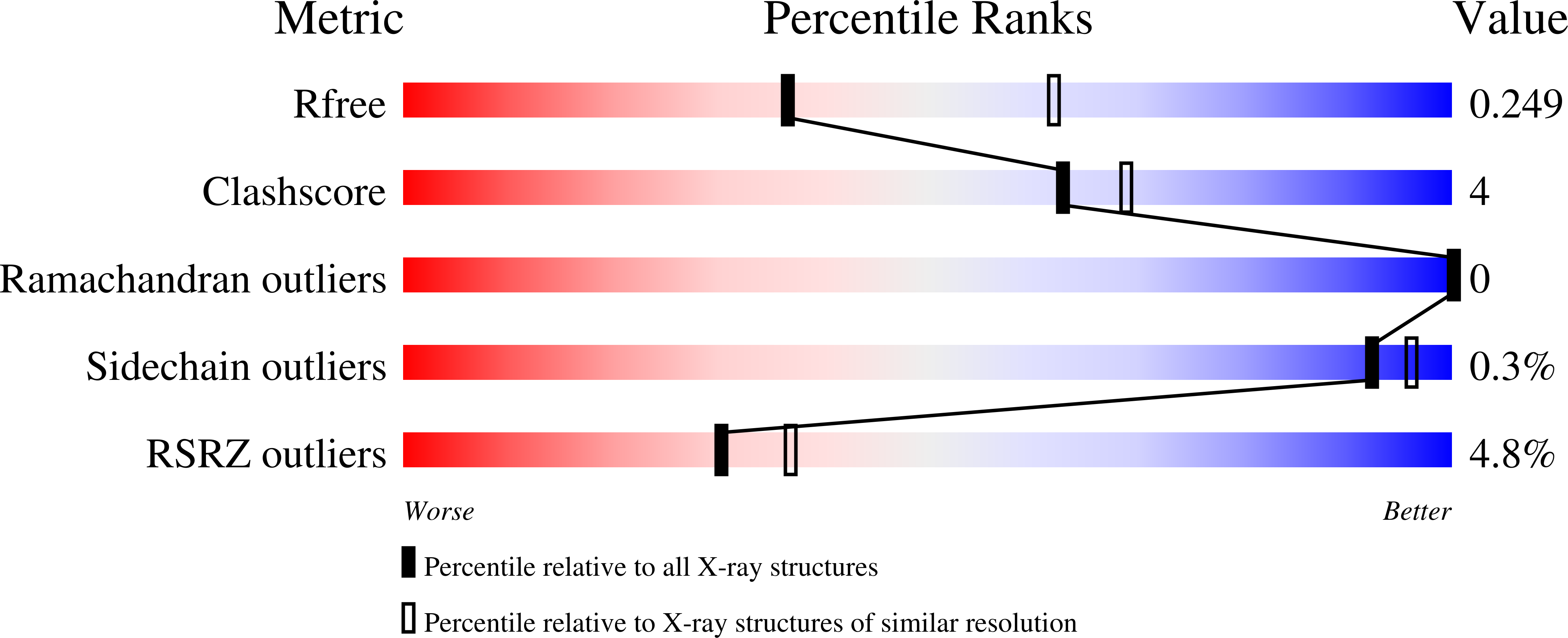
5TGS, 5TH5 - PubMed Abstract:
Radical S-adenosyl-l-methionine (SAM) enzymes are widely distributed and catalyze diverse reactions. SAM binds to the unique iron atom of a site-differentiated [4Fe-4S] cluster and is reductively cleaved to generate a 5'-deoxyadenosyl radical, which initiates turnover. 7-Carboxy-7-deazaguanine (CDG) synthase (QueE) catalyzes a key step in the biosynthesis of 7-deazapurine containing natural products. 6-Carboxypterin (6-CP), an oxidized analogue of the natural substrate 6-carboxy-5,6,7,8-tetrahydropterin (CPH 4 ), is shown to be an alternate substrate for CDG synthase. Under reducing conditions that would promote the reductive cleavage of SAM, 6-CP is turned over to 6-deoxyadenosylpterin (6-dAP), presumably by radical addition of the 5'-deoxyadenosine followed by oxidative decarboxylation to the product. By contrast, in the absence of the strong reductant, dithionite, the carboxylate of 6-CP is esterified to generate 6-carboxypterin-5'-deoxyadenosyl ester (6-CP-dAdo ester). Structural studies with 6-CP and SAM also reveal electron density consistent with the ester product being formed in crystallo. The differential reactivity of 6-CP under reducing and nonreducing conditions highlights the ability of radical SAM enzymes to carry out both polar and radical transformations in the same active site.
Organizational Affiliation:
Department of Chemistry, University of Utah , Salt Lake City, Utah 84112, United States.