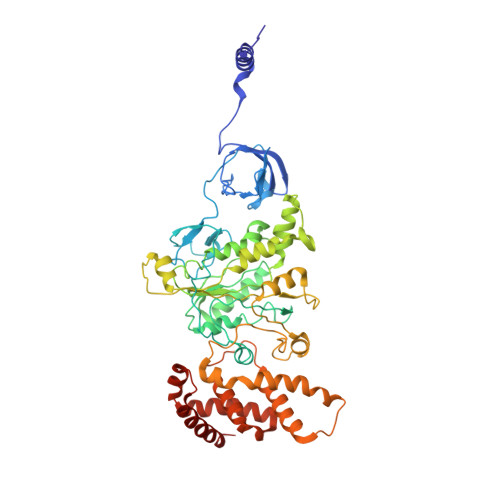
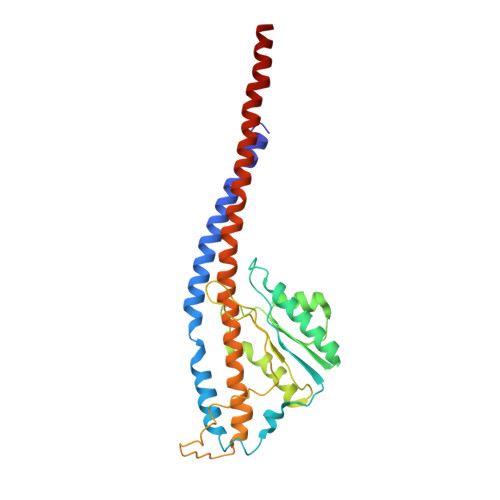
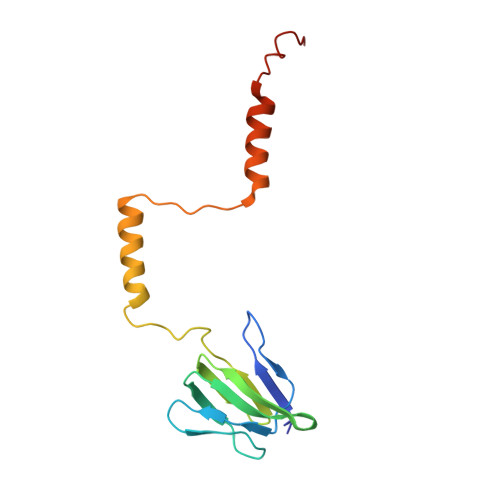
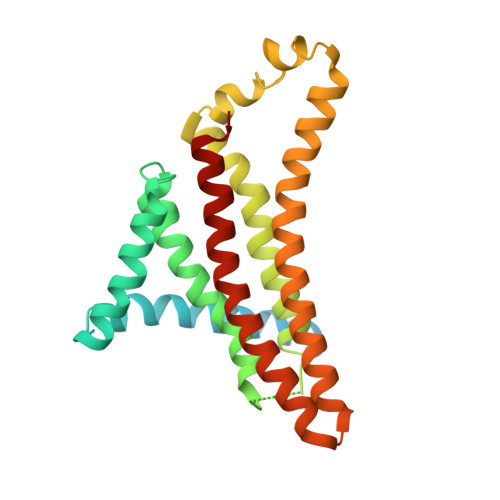
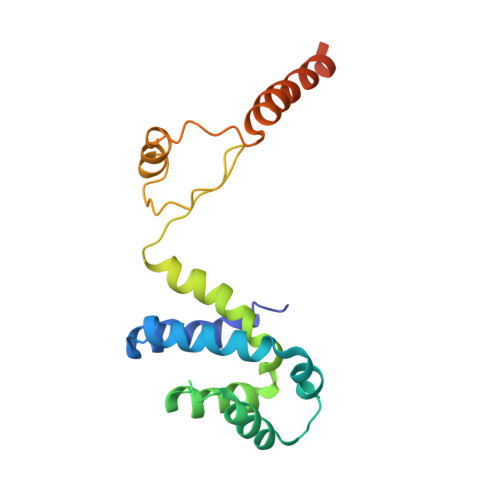
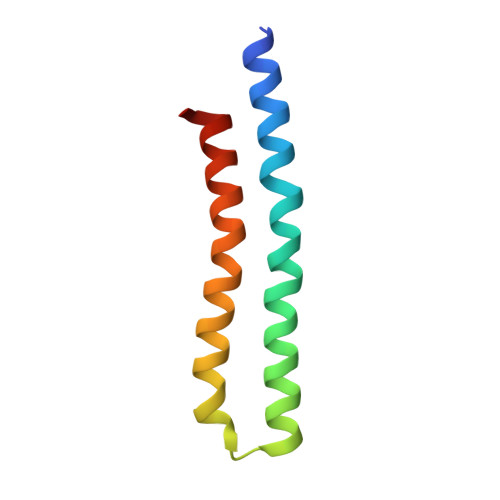
Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Sobti, M., Smits, C., Wong, A.S., Ishmukhametov, R., Stock, D., Sandin, S., Stewart, A.G.(2016) Elife 5
- PubMed: 28001127
- DOI: https://doi.org/10.7554/eLife.21598
- Primary Citation of Related Structures:
5T4O, 5T4P, 5T4Q - PubMed Abstract:
A molecular model that provides a framework for interpreting the wealth of functional information obtained on the E. coli F-ATP synthase has been generated using cryo-electron microscopy. Three different states that relate to rotation of the enzyme were observed, with the central stalk's ε subunit in an extended autoinhibitory conformation in all three states. The F o motor comprises of seven transmembrane helices and a decameric c-ring and invaginations on either side of the membrane indicate the entry and exit channels for protons. The proton translocating subunit contains near parallel helices inclined by ~30° to the membrane, a feature now synonymous with rotary ATPases. For the first time in this rotary ATPase subtype, the peripheral stalk is resolved over its entire length of the complex, revealing the F 1 attachment points and a coiled-coil that bifurcates toward the membrane with its helices separating to embrace subunit a from two sides.
Organizational Affiliation:
Molecular, Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Darlinghurst, Australia.