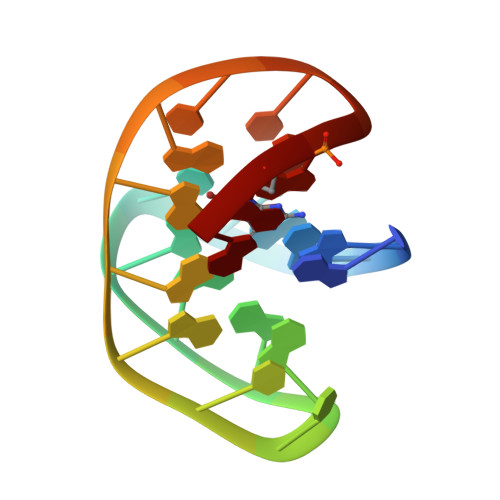
NMR structure of a G-quadruplex formed by four d(G4C2) repeats: insights into structural polymorphism.
Brcic, J., Plavec, J.(2018) Nucleic Acids Res 46: 11605-11617
- PubMed: 30277522
- DOI: https://doi.org/10.1093/nar/gky886
- Primary Citation of Related Structures:
5OPH - PubMed Abstract:
Most frequent genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), is a largely increased number of d(G4C2)n•(G2C4)n repeats located in the non-coding region of C9orf72 gene. Non-canonical structures, including G-quadruplexes, formed within expanded repeats have been proposed to drive repeat expansion and pathogenesis of ALS and FTD. Oligonucleotide d[(G4C2)3G4], which represents the shortest oligonucleotide model of d(G4C2) repeats with the ability to form a unimolecular G-quadruplex, forms two major G-quadruplex structures in addition to several minor species which coexist in solution with K+ ions. Herein, we used solution-state NMR to determine the high-resolution structure of one of the major G-quadruplex species adopted by d[(G4C2)3G4]. Structural characterization of the G-quadruplex named AQU was facilitated by a single substitution of dG with 8Br-dG at position 21 and revealed an antiparallel fold composed of four G-quartets and three lateral C-C loops. The G-quadruplex exhibits high thermal stability and is favored kinetically and under slightly acidic conditions. An unusual structural element distinct from a C-quartet is observed in the structure. Two C•C base pairs are stacked on the nearby G-quartet and are involved in a dynamic equilibrium between symmetric N3-amino and carbonyl-amino geometries and protonated C+•C state.
Organizational Affiliation:
Slovenian NMR Center, National Institute of Chemistry, Ljubljana SI-1000, Slovenia.