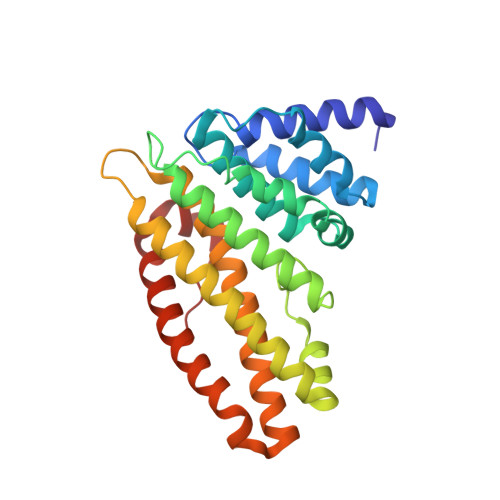
Epsin and Sla2 form assemblies through phospholipid interfaces.
Garcia-Alai, M.M., Heidemann, J., Skruzny, M., Gieras, A., Mertens, H.D.T., Svergun, D.I., Kaksonen, M., Uetrecht, C., Meijers, R.(2018) Nat Commun 9: 328-328
- PubMed: 29362354
- DOI: https://doi.org/10.1038/s41467-017-02443-x
- Primary Citation of Related Structures:
5ON7, 5ONF, 5OO7, 6ENR - PubMed Abstract:
In clathrin-mediated endocytosis, adapter proteins assemble together with clathrin through interactions with specific lipids on the plasma membrane. However, the precise mechanism of adapter protein assembly at the cell membrane is still unknown. Here, we show that the membrane-proximal domains ENTH of epsin and ANTH of Sla2 form complexes through phosphatidylinositol 4,5-bisphosphate (PIP2) lipid interfaces. Native mass spectrometry reveals how ENTH and ANTH domains form assemblies by sharing PIP2 molecules. Furthermore, crystal structures of epsin Ent2 ENTH domain from S. cerevisiae in complex with PIP2 and Sla2 ANTH domain from C. thermophilum illustrate how allosteric phospholipid binding occurs. A comparison with human ENTH and ANTH domains reveal only the human ENTH domain can form a stable hexameric core in presence of PIP2, which could explain functional differences between fungal and human epsins. We propose a general phospholipid-driven multifaceted assembly mechanism tolerating different adapter protein compositions to induce endocytosis.
Organizational Affiliation:
European Molecular Biology Laboratory (EMBL), Hamburg Outstation, Notkestrasse 85, 22607, Hamburg, Germany.