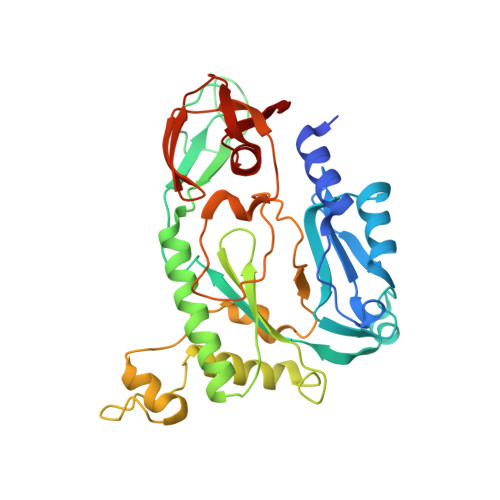
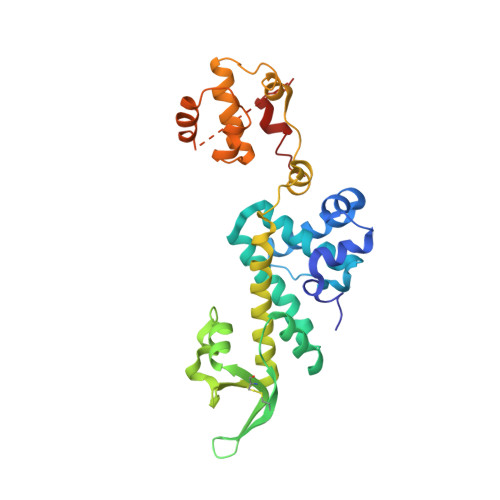
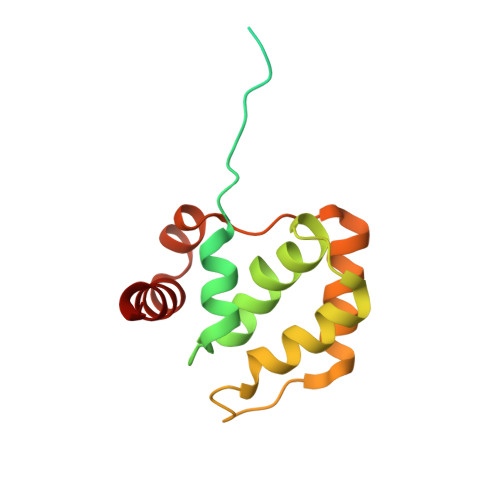
Primer synthesis by a eukaryotic-like archaeal primase is independent of its Fe-S cluster.
Holzer, S., Yan, J., Kilkenny, M.L., Bell, S.D., Pellegrini, L.(2017) Nat Commun 8: 1718-1718
- PubMed: 29167441
- DOI: https://doi.org/10.1038/s41467-017-01707-w
- Primary Citation of Related Structures:
5OF3, 5OFN - PubMed Abstract:
DNA replication depends on primase, the specialised polymerase responsible for synthesis of the RNA primers that are elongated by the replicative DNA polymerases. In eukaryotic and archaeal replication, primase is a heterodimer of two subunits, PriS and PriL. Recently, a third primase subunit named PriX was identified in the archaeon Sulfolobus solfataricus. PriX is essential for primer synthesis and is structurally related to the Fe-S cluster domain of eukaryotic PriL. Here we show that PriX contains a nucleotide-binding site required for primer synthesis, and demonstrate equivalence of nucleotide-binding residues in PriX with eukaryotic PriL residues that are known to be important for primer synthesis. A primase chimera, where PriX is fused to a truncated version of PriL lacking the Fe-S cluster domain retains wild-type levels of primer synthesis. Our evidence shows that PriX has replaced PriL as the subunit that endows primase with the unique ability to initiate nucleic acid synthesis. Importantly, our findings reveal that the Fe-S cluster is not required for primer synthesis.
- Department of Biochemistry, University of Cambridge, Tennis Court Road, Cambridge, UK.
Organizational Affiliation: