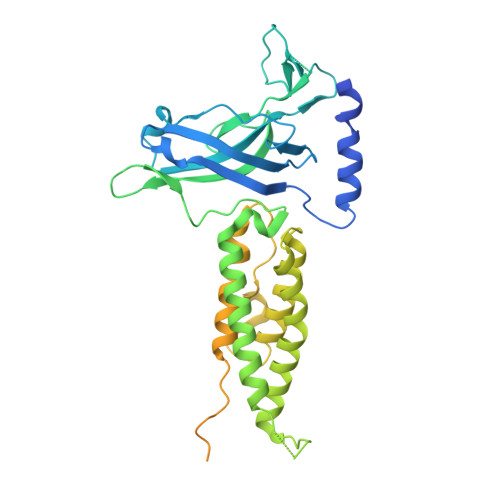
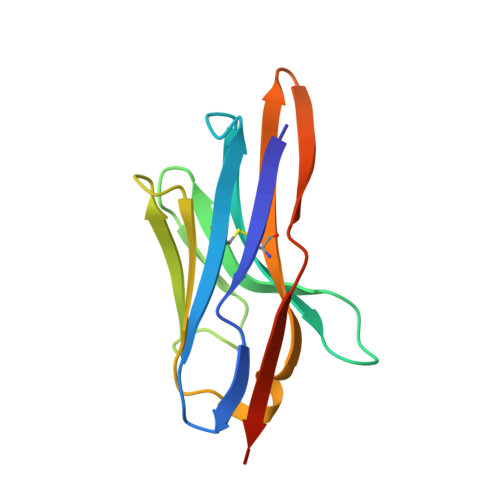
Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nguyen, V.S., Logger, L., Spinelli, S., Legrand, P., Huyen Pham, T.T., Nhung Trinh, T.T., Cherrak, Y., Zoued, A., Desmyter, A., Durand, E., Roussel, A., Kellenberger, C., Cascales, E., Cambillau, C.(2017) Nat Microbiol 2: 17103-17103
- PubMed: 28650463
- DOI: https://doi.org/10.1038/nmicrobiol.2017.103
- Primary Citation of Related Structures:
5M2W, 5M2Y, 5M30, 5MWN - PubMed Abstract:
The type VI secretion system (T6SS) is a multiprotein machine widespread in Gram-negative bacteria that delivers toxins into both eukaryotic and prokaryotic cells. The mechanism of action of the T6SS is comparable to that of contractile myophages. The T6SS builds a tail-like structure made of an inner tube wrapped by a sheath, assembled under an extended conformation. Contraction of the sheath propels the inner tube towards the target cell. The T6SS tail is assembled on a platform-the baseplate-which is functionally similar to bacteriophage baseplates. In addition, the baseplate docks the tail to a trans-envelope membrane complex that orients the tail towards the target. Here, we report the crystal structure of TssK, a central component of the T6SS baseplate. We show that TssK is composed of three domains, and establish the contribution of each domain to the interaction with TssK partners. Importantly, this study reveals that the N-terminal domain of TssK is structurally homologous to the shoulder domain of phage receptor-binding proteins, and the C-terminal domain binds the membrane complex. We propose that TssK has conserved the domain of attachment to the virion particle but has evolved the reception domain to use the T6SS membrane complex as receptor.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, Aix-Marseille Université, Campus de Luminy, Case 932, 13288 Marseille Cedex 09, France.