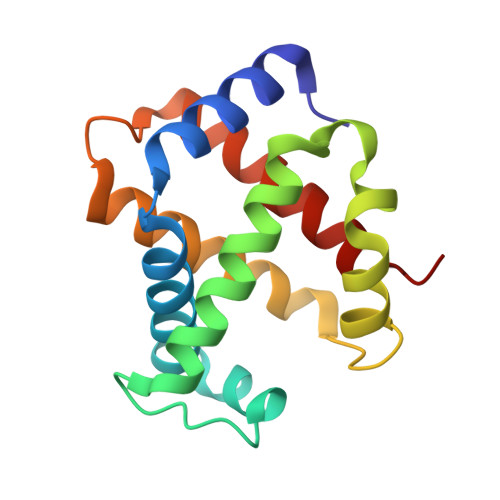
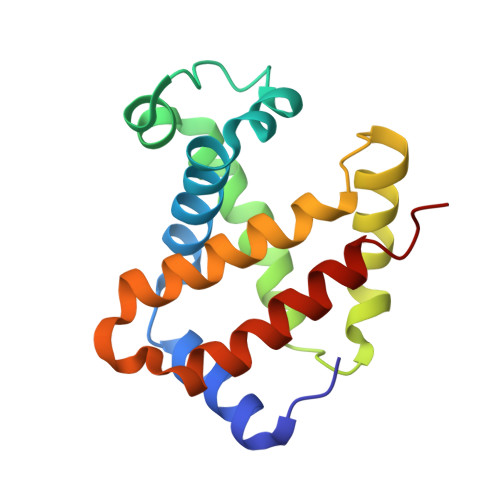
Alteration of the alpha 1 beta 2/ alpha 2 beta 1 subunit interface contributes to the increased hemoglobin-oxygen affinity of high-altitude deer mice.
Inoguchi, N., Mizuno, N., Baba, S., Kumasaka, T., Natarajan, C., Storz, J.F., Moriyama, H.(2017) PLoS One 12: e0174921-e0174921
- PubMed: 28362841
- DOI: https://doi.org/10.1371/journal.pone.0174921
- Primary Citation of Related Structures:
5KER - PubMed Abstract:
Deer mice (Peromyscus maniculatus) that are native to high altitudes in the Rocky Mountains have evolved hemoglobins with an increased oxygen-binding affinity relative to those of lowland conspecifics. To elucidate the molecular mechanisms responsible for the evolved increase in hemoglobin-oxygen affinity, the crystal structure of the highland hemoglobin variant was solved and compared with the previously reported structure for the lowland variant. Highland hemoglobin yielded at least two crystal types, in which the longest axes were 507 and 230 Å. Using the smaller unit cell crystal, the structure was solved at 2.2 Å resolution. The asymmetric unit contained two tetrameric hemoglobin molecules. The analyses revealed that αPro50 in the highland hemoglobin variant promoted a stable interaction between αHis45 and heme that was not seen in the αHis50 lowland variant. The αPro50 mutation also altered the nature of atomic contacts at the α1β2/α2β1 intersubunit interfaces. These results demonstrate how affinity-altering changes in intersubunit interactions can be produced by mutations at structurally remote sites.
Organizational Affiliation:
School of Biological Sciences, University of Nebraska, Lincoln, Nebraska, United States of America.