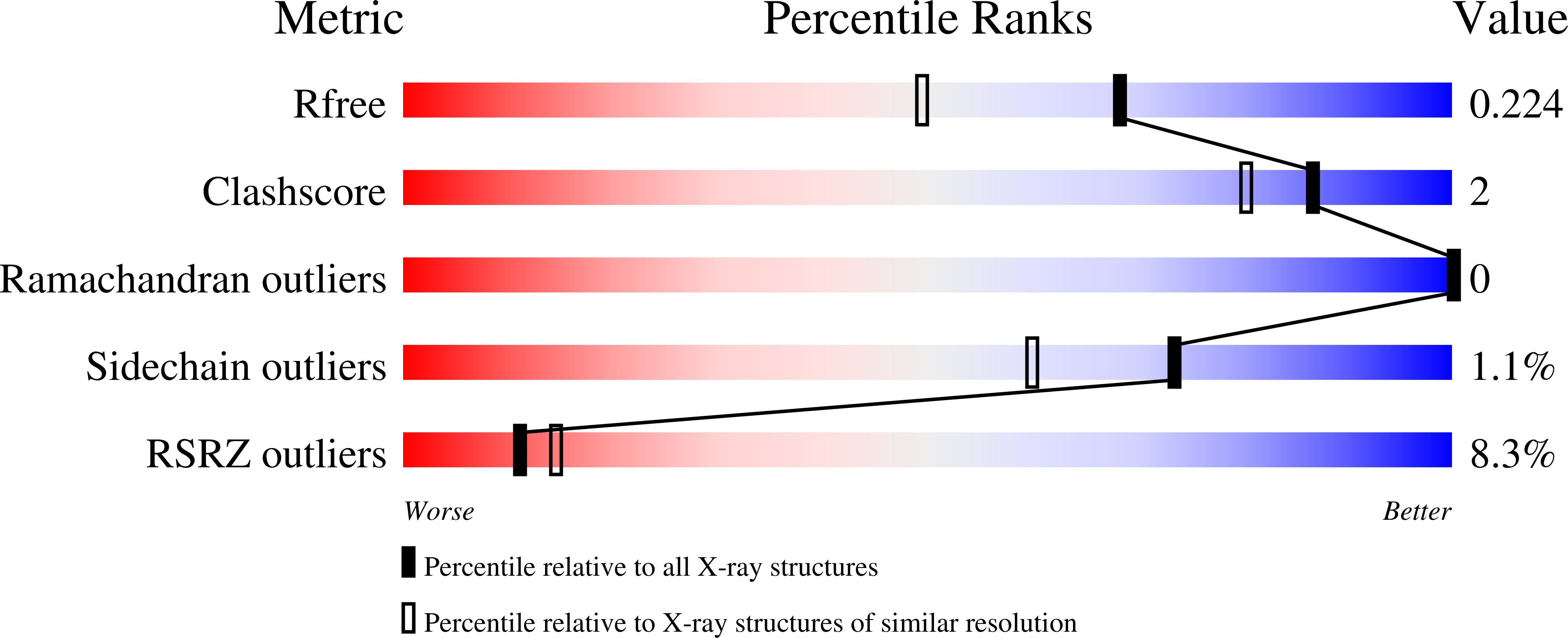
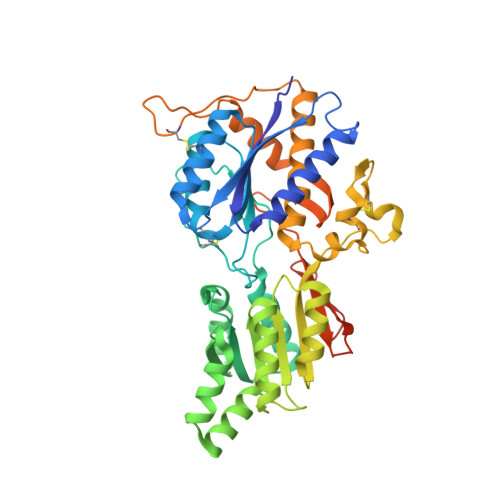
Structural basis for integration of GluD receptors within synaptic organizer complexes.
Elegheert, J., Kakegawa, W., Clay, J.E., Shanks, N.F., Behiels, E., Matsuda, K., Kohda, K., Miura, E., Rossmann, M., Mitakidis, N., Motohashi, J., Chang, V.T., Siebold, C., Greger, I.H., Nakagawa, T., Yuzaki, M., Aricescu, A.R.(2016) Science 353: 295-299
- PubMed: 27418511
- DOI: https://doi.org/10.1126/science.aae0104
- Primary Citation of Related Structures:
5KC5, 5KC6, 5KC7, 5KC8, 5KC9, 5KCA - PubMed Abstract:
Ionotropic glutamate receptor (iGluR) family members are integrated into supramolecular complexes that modulate their location and function at excitatory synapses. However, a lack of structural information beyond isolated receptors or fragments thereof currently limits the mechanistic understanding of physiological iGluR signaling. Here, we report structural and functional analyses of the prototypical molecular bridge linking postsynaptic iGluR δ2 (GluD2) and presynaptic β-neurexin 1 (β-NRX1) via Cbln1, a C1q-like synaptic organizer. We show how Cbln1 hexamers "anchor" GluD2 amino-terminal domain dimers to monomeric β-NRX1. This arrangement promotes synaptogenesis and is essential for D: -serine-dependent GluD2 signaling in vivo, which underlies long-term depression of cerebellar parallel fiber-Purkinje cell (PF-PC) synapses and motor coordination in developing mice. These results lead to a model where protein and small-molecule ligands synergistically control synaptic iGluR function.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.