Inactivation of Urease by Catechol: Kinetics and Structure.
Mazzei, L., Cianci, M., Musiani, F., Lente, G., Palombo, M., Ciurli, S.(2016) J Inorg Biochem 166: 182
- PubMed: 27888701
- DOI: https://doi.org/10.1016/j.jinorgbio.2016.11.016
- Primary Citation of Related Structures:
5G4H - PubMed Abstract:
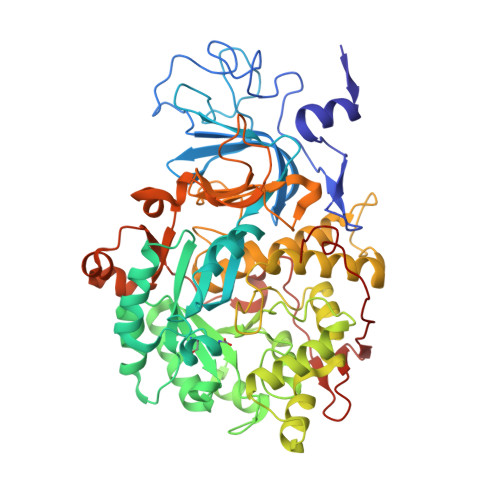
Urease is a Ni(II)-containing enzyme that catalyzes the hydrolysis of urea to yield ammonia and carbamate at a rate 10 15 times higher than the uncatalyzed reaction. Urease is a virulence factor of several human pathogens, in addition to decreasing the efficiency of soil organic nitrogen fertilization. Therefore, efficient urease inhibitors are actively sought. In this study, we describe a molecular characterization of the interaction between urease from Sporosarcina pasteurii (SPU) and Canavalia ensiformis (jack bean, JBU) with catechol, a model polyphenol. In particular, catechol irreversibly inactivates both SPU and JBU with a complex radical-based autocatalytic multistep mechanism. The crystal structure of the SPU-catechol complex, determined at 1.50Å resolution, reveals the structural details of the enzyme inhibition.
Organizational Affiliation:
Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Italy.