Contribution of Physical Interactions to Signaling Specificity between a Diguanylate Cyclase and Its Effector.
Dahlstrom, K.M., Giglio, K.M., Collins, A.J., Sondermann, H., O'Toole, G.A.(2015) mBio 6: e01978-e01915
- PubMed: 26670387
- DOI: https://doi.org/10.1128/mBio.01978-15
- Primary Citation of Related Structures:
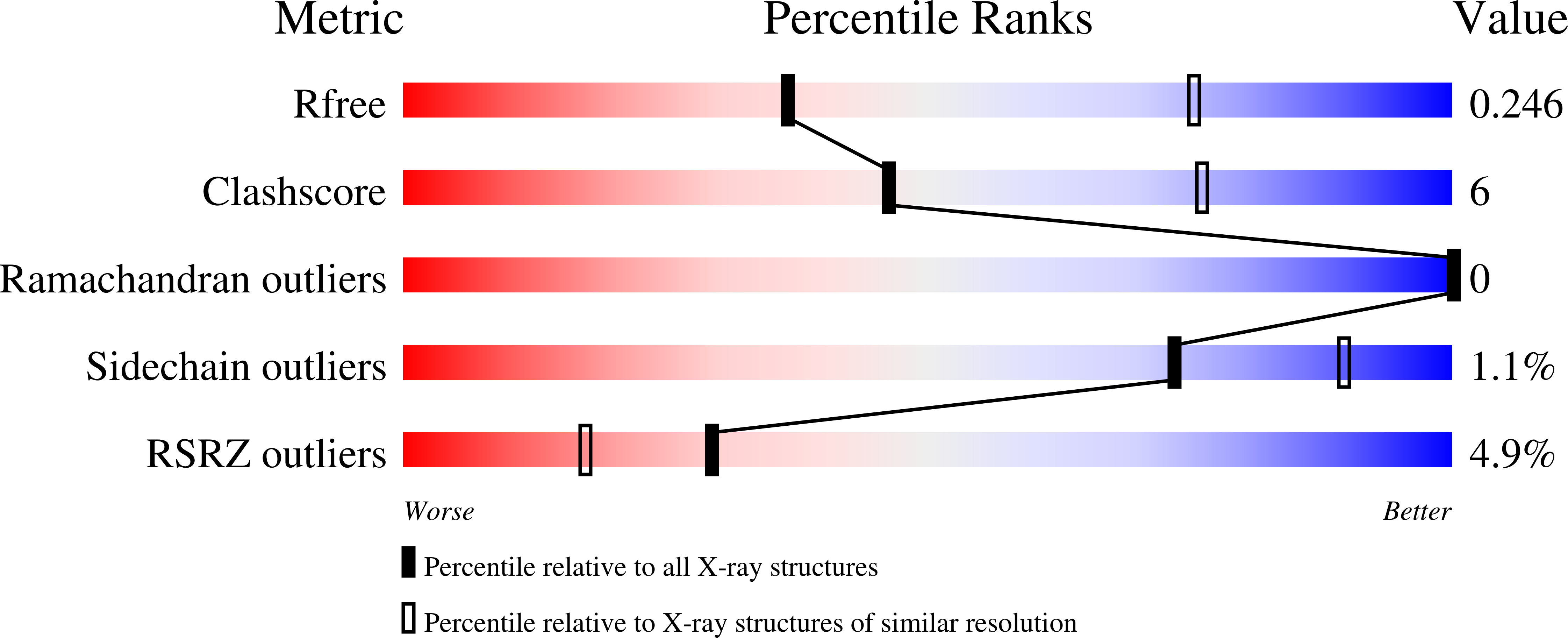
5EUH - PubMed Abstract:
Cyclic diguanylate (c-di-GMP) is a bacterial second messenger that controls multiple cellular processes. c-di-GMP networks have up to dozens of diguanylate cyclases (DGCs) that synthesize c-di-GMP along with many c-di-GMP-responsive target proteins that can bind and respond to this signal. For such networks to have order, a mechanism(s) likely exists that allow DGCs to specifically signal their targets, and it has been suggested that physical interactions might provide such specificity. Our results show a DGC from Pseudomonas fluorescens physically interacting with its target protein at a conserved interface, and this interface can be predictive of DGC-target protein interactions. Furthermore, we demonstrate that physical interaction is necessary for the DGC to maximally signal its target. If such "local signaling" is a theme for even a fraction of the DGCs used by bacteria, it becomes possible to posit a model whereby physical interaction allows a DGC to directly signal its target protein, which in turn may help curtail undesired cross talk with other members of the network. An important question in microbiology is how bacteria make decisions using a signaling network made up of proteins that make, break, and bind the second messenger c-di-GMP, which is responsible for controlling many cellular behaviors. Previous work has shown that a given DGC enzyme will signal for specific cellular outputs, despite making the same diffusible molecule as its sibling DGCs in the unpartitioned space of the bacterial cell. Understanding how one DGC differentiates its output from the dozens of other such enzymes in the cell is synonymous with understanding a large component of the bacterial decision-making machinery. We present evidence for a helix on a DGC used to physically associate with its target protein, which is necessary to achieve maximal signaling.
Organizational Affiliation:
Department of Microbiology and Immunology, Geisel School of Medicine at Dartmouth, Hanover, New Hampshire.