Functional Dissection of the Bipartite Active Site of the Class I Coenzyme A (CoA)-Transferase Succinyl-CoA:Acetate CoA-Transferase.
Murphy, J.R., Mullins, E.A., Kappock, T.J.(2016) Front Chem 4: 23-23
- PubMed: 27242998
- DOI: https://doi.org/10.3389/fchem.2016.00023
- Primary Citation of Related Structures:
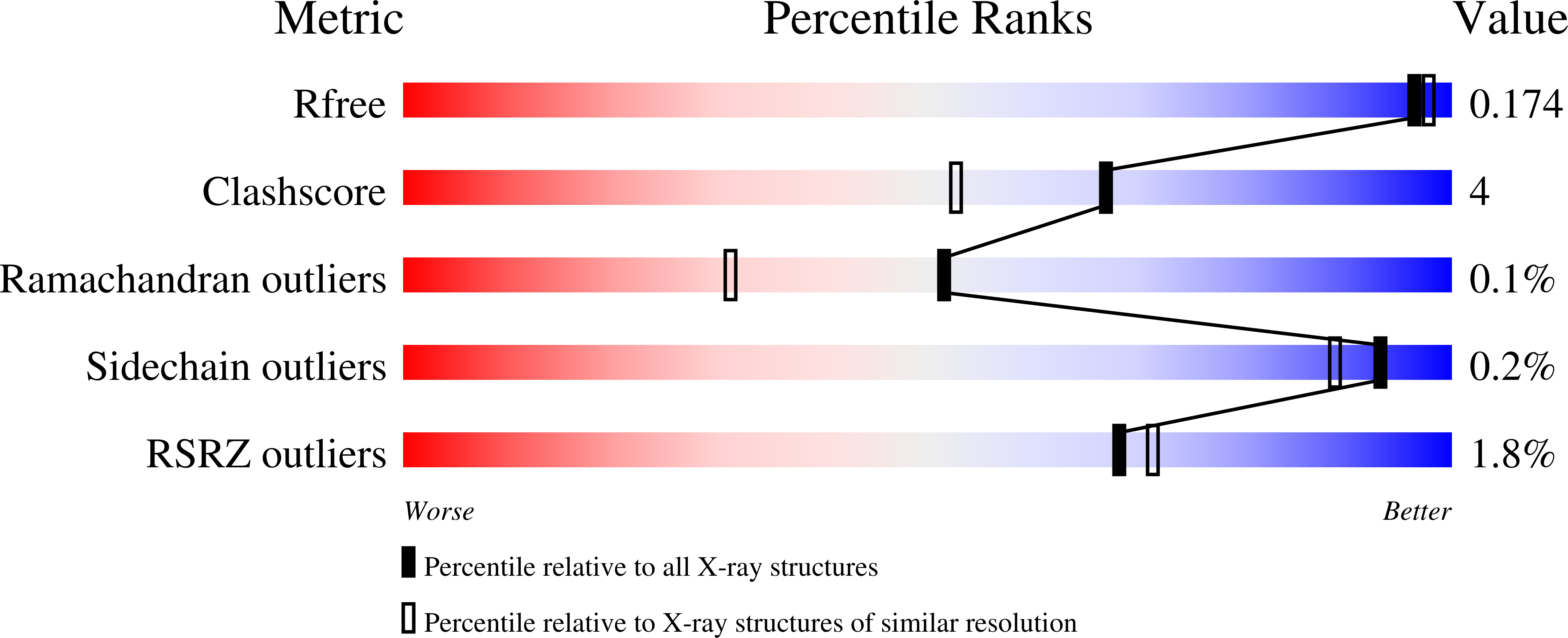
5DDK, 5DW4, 5DW5, 5DW6, 5E5H - PubMed Abstract:
Coenzyme A (CoA)-transferases catalyze the reversible transfer of CoA from acyl-CoA thioesters to free carboxylates. Class I CoA-transferases produce acylglutamyl anhydride intermediates that undergo attack by CoA thiolate on either the internal or external carbonyl carbon atoms, forming distinct tetrahedral intermediates <3 Å apart. In this study, crystal structures of succinyl-CoA:acetate CoA-transferase (AarC) from Acetobacter aceti are used to examine how the Asn347 carboxamide stabilizes the internal oxyanion intermediate. A structure of the active mutant AarC-N347A bound to CoA revealed both solvent replacement of the missing contact and displacement of the adjacent Glu294, indicating that Asn347 both polarizes and orients the essential glutamate. AarC was crystallized with the nonhydrolyzable acetyl-CoA (AcCoA) analog dethiaacetyl-CoA (1a) in an attempt to trap a closed enzyme complex containing a stable analog of the external oxyanion intermediate. One active site contained an acetylglutamyl anhydride adduct and truncated 1a, an unexpected result hinting at an unprecedented cleavage of the ketone moiety in 1a. Solution studies confirmed that 1a decomposition is accompanied by production of near-stoichiometric acetate, in a process that seems to depend on microbial contamination but not AarC. A crystal structure of AarC bound to the postulated 1a truncation product (2a) showed complete closure of one active site per dimer but no acetylglutamyl anhydride, even with acetate added. These findings suggest that an activated acetyl donor forms during 1a decomposition; a working hypothesis involving ketone oxidation is offered. The ability of 2a to induce full active site closure furthermore suggests that it subverts a system used to impede inappropriate active site closure on unacylated CoA.
Organizational Affiliation:
Department of Biochemistry, Purdue University West Lafayette, IN, USA.