New IDH1 mutant inhibitors for treatment of acute myeloid leukemia.
Okoye-Okafor, U.C., Bartholdy, B., Cartier, J., Gao, E.N., Pietrak, B., Rendina, A.R., Rominger, C., Quinn, C., Smallwood, A., Wiggall, K.J., Reif, A.J., Schmidt, S.J., Qi, H., Zhao, H., Joberty, G., Faelth-Savitski, M., Bantscheff, M., Drewes, G., Duraiswami, C., Brady, P., Groy, A., Narayanagari, S.R., Antony-Debre, I., Mitchell, K., Wang, H.R., Kao, Y.R., Christopeit, M., Carvajal, L., Barreyro, L., Paietta, E., Makishima, H., Will, B., Concha, N., Adams, N.D., Schwartz, B., McCabe, M.T., Maciejewski, J., Verma, A., Steidl, U.(2015) Nat Chem Biol 11: 878-886
- PubMed: 26436839
- DOI: https://doi.org/10.1038/nchembio.1930
- Primary Citation of Related Structures:
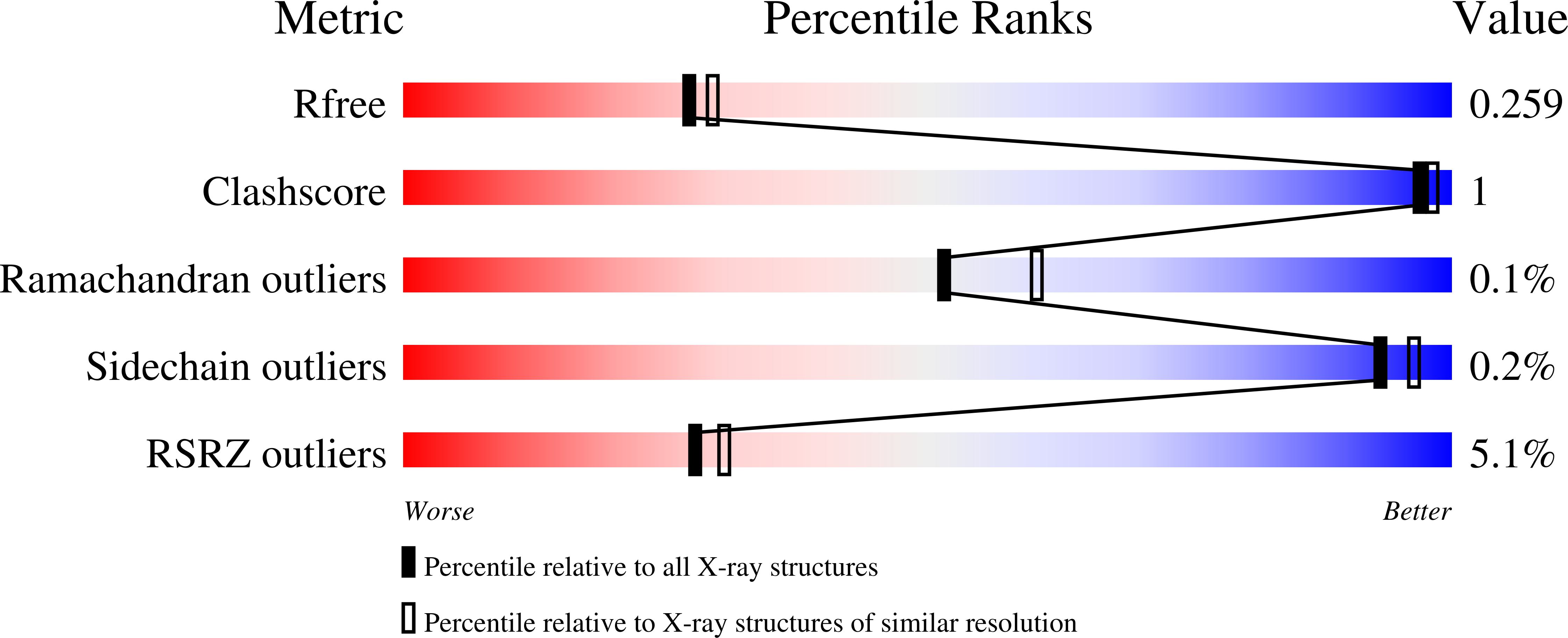
5DE1 - PubMed Abstract:
Neomorphic mutations in isocitrate dehydrogenase 1 (IDH1) are driver mutations in acute myeloid leukemia (AML) and other cancers. We report the development of new allosteric inhibitors of mutant IDH1. Crystallographic and biochemical results demonstrated that compounds of this chemical series bind to an allosteric site and lock the enzyme in a catalytically inactive conformation, thereby enabling inhibition of different clinically relevant IDH1 mutants. Treatment of IDH1 mutant primary AML cells uniformly led to a decrease in intracellular 2-HG, abrogation of the myeloid differentiation block and induction of granulocytic differentiation at the level of leukemic blasts and more immature stem-like cells, in vitro and in vivo. Molecularly, treatment with the inhibitors led to a reversal of the DNA cytosine hypermethylation patterns caused by mutant IDH1 in the cells of individuals with AML. Our study provides proof of concept for the molecular and biological activity of novel allosteric inhibitors for targeting different mutant forms of IDH1 in leukemia.
Organizational Affiliation:
Department of Cell Biology, Albert Einstein College of Medicine, Bronx, New York, USA.