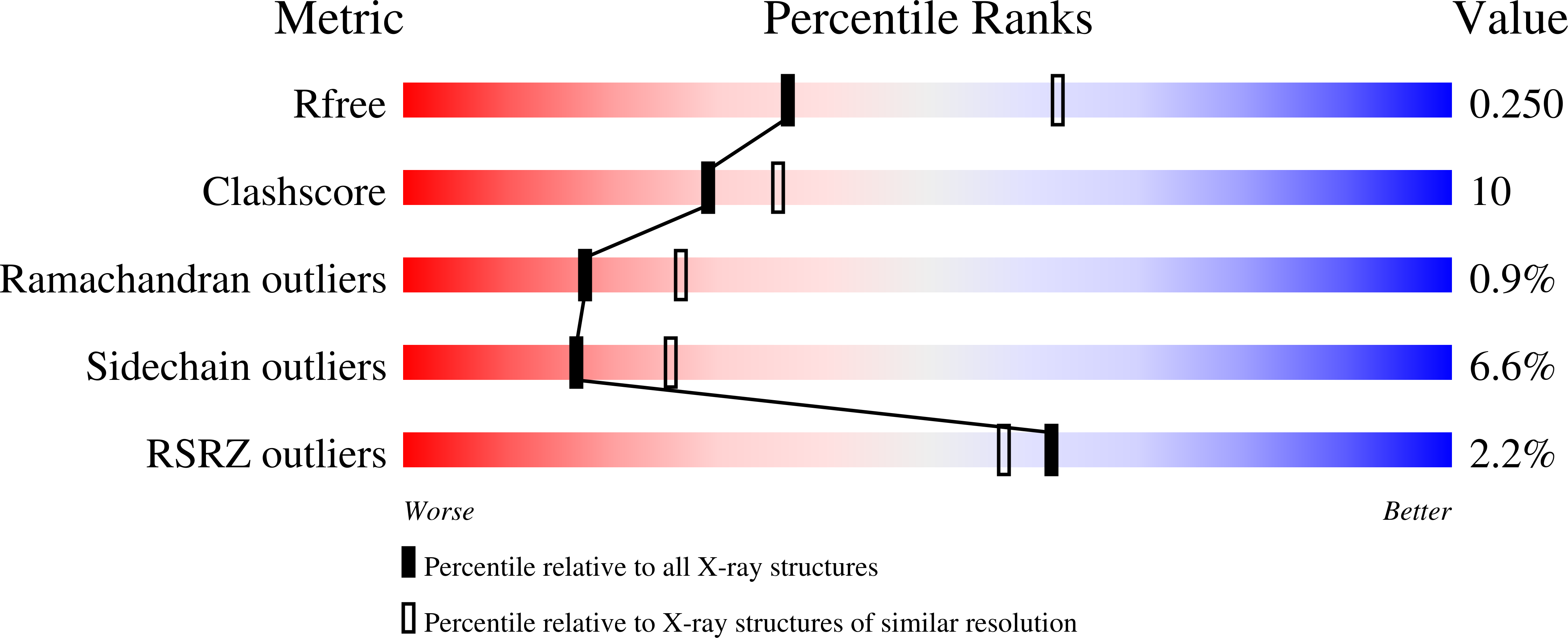
Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.
Payne, K.A., Fisher, K., Sjuts, H., Dunstan, M.S., Bellina, B., Johannissen, L., Barran, P., Hay, S., Rigby, S.E., Leys, D.(2015) J Biol Chem 290: 27572-27581
- PubMed: 26378237
- DOI: https://doi.org/10.1074/jbc.M115.685693
- Primary Citation of Related Structures:
5D6S - PubMed Abstract:
Queuosine (Q) is a hypermodified RNA base that replaces guanine in the wobble positions of 5'-GUN-3' tRNA molecules. Q is exclusively made by bacteria, and the corresponding queuine base is a micronutrient salvaged by eukaryotic species. The final step in Q biosynthesis is the reduction of the epoxide precursor, epoxyqueuosine, to yield the Q cyclopentene ring. The epoxyqueuosine reductase responsible, QueG, shares distant homology with the cobalamin-dependent reductive dehalogenase (RdhA), however the role played by cobalamin in QueG catalysis has remained elusive. We report the solution and structural characterization of Streptococcus thermophilus QueG, revealing the enzyme harbors a redox chain consisting of two [4Fe-4S] clusters and a cob(II)alamin in the base-off form, similar to RdhAs. In contrast to the shared redox chain architecture, the QueG active site shares little homology with RdhA, with the notable exception of a conserved Tyr that is proposed to function as a proton donor during reductive dehalogenation. Docking of an epoxyqueuosine substrate suggests the QueG active site places the substrate cyclopentane moiety in close proximity of the cobalt. Both the Tyr and a conserved Asp are implicated as proton donors to the epoxide leaving group. This suggests that, in contrast to the unusual carbon-halogen bond chemistry catalyzed by RdhAs, QueG acts via Co-C bond formation. Our study establishes the common features of Class III cobalamin-dependent enzymes, and reveals an unexpected diversity in the reductive chemistry catalyzed by these enzymes.
Organizational Affiliation:
From the Manchester Institute of Biotechnology, University of Manchester, Princess Street 131, Manchester M1 7DN, United Kingdom karl.payne@manchester.ac.uk.