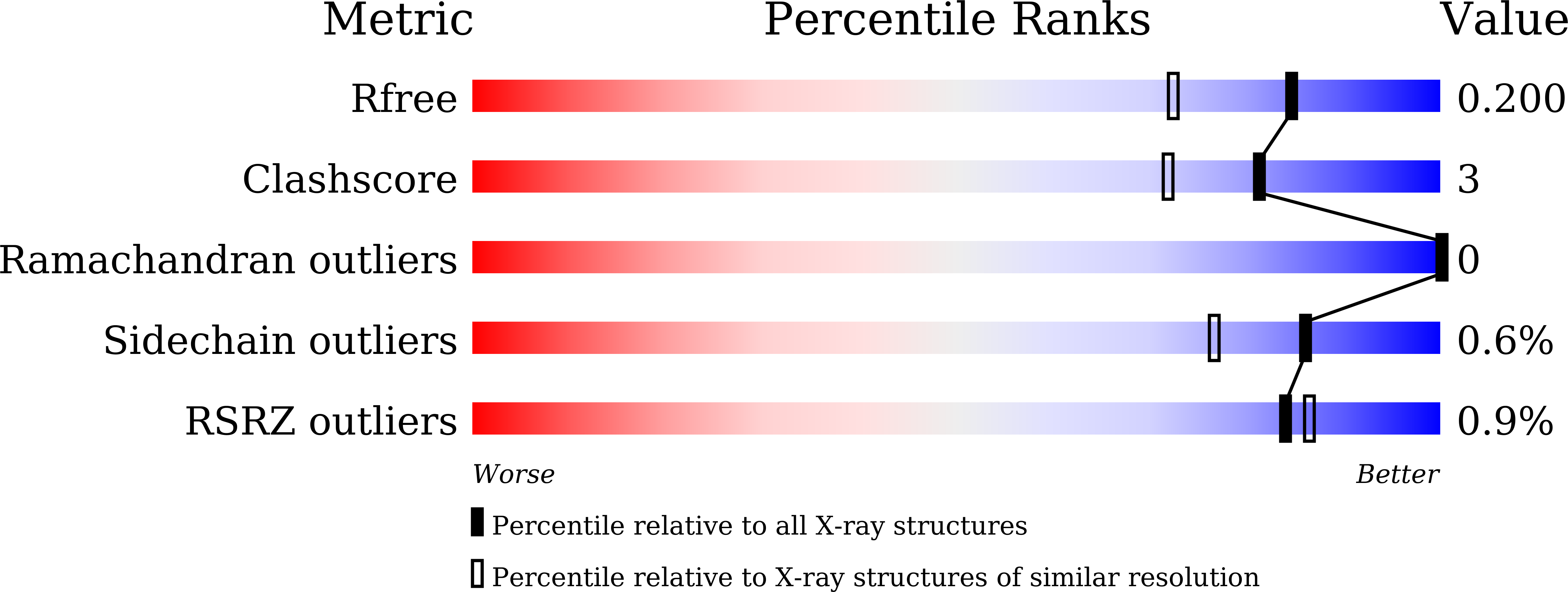
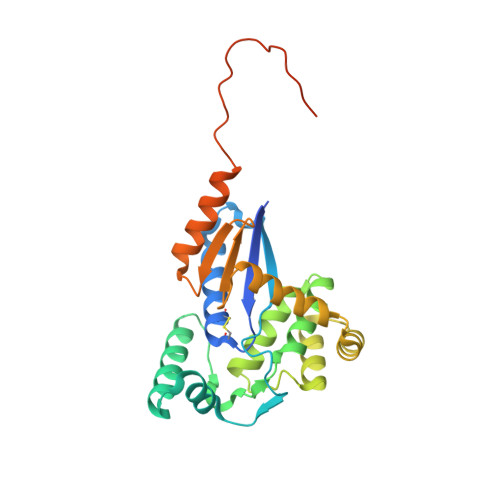
drFrnE Represents a Hitherto Unknown Class of Eubacterial Cytoplasmic Disulfide Oxido-Reductases.
Bihani, S.C., Panicker, L., Rajpurohit, Y.S., Misra, H.S., Kumar, V.(2018) Antioxid Redox Signal 28: 296-310
- PubMed: 28899103
- DOI: https://doi.org/10.1089/ars.2016.6960
- Primary Citation of Related Structures:
5CNW, 5CO3, 5COH, 5E59 - PubMed Abstract:
Living cells employ thioredoxin and glutaredoxin disulfide oxido-reductases to protect thiol groups in intracellular proteins. FrnE protein of Deinococcus radiodurans (drFrnE) is a disulfide oxido-reductase that is induced in response to Cd 2+ exposure and is involved in cadmium and radiation tolerance. The aim of this study is to probe structure, function, and cellular localization of FrnE class of proteins.
Organizational Affiliation:
1 Protein Crystallography Section, Radiation Biology & Health Sciences Division, Bhabha Atomic Research Centre , Mumbai, India .