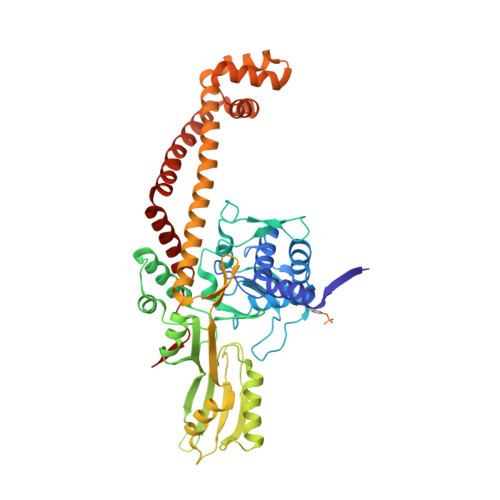
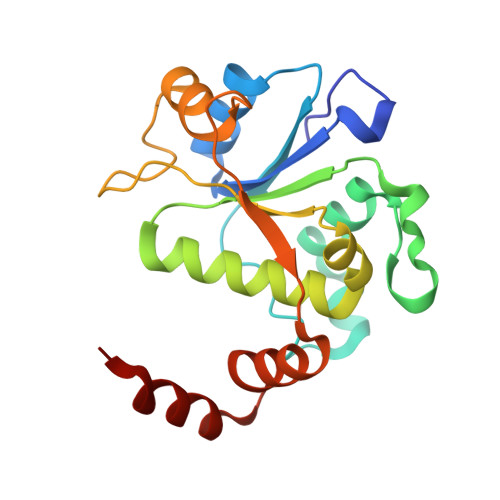
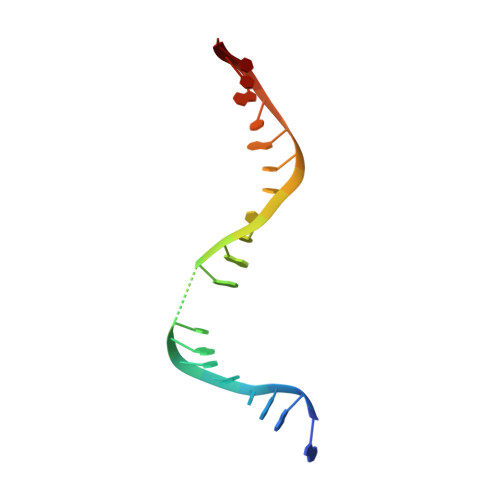
Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Chan, P.F., Srikannathasan, V., Huang, J., Cui, H., Fosberry, A.P., Gu, M., Hann, M.M., Hibbs, M., Homes, P., Ingraham, K., Pizzollo, J., Shen, C., Shillings, A.J., Spitzfaden, C.E., Tanner, R., Theobald, A.J., Stavenger, R.A., Bax, B.D., Gwynn, M.N.(2015) Nat Commun 6: 10048-10048
- PubMed: 26640131
- DOI: https://doi.org/10.1038/ncomms10048
- Primary Citation of Related Structures:
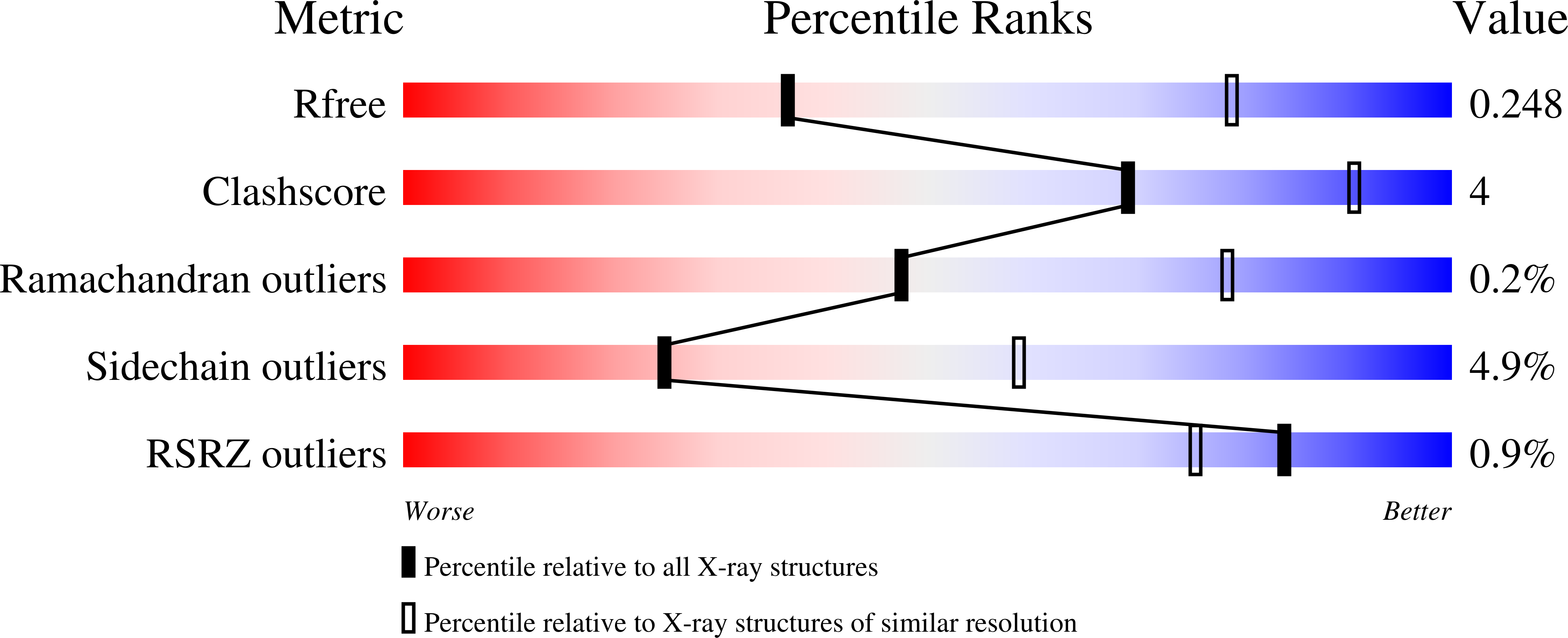
5CDM, 5CDN, 5CDO, 5CDP, 5CDQ, 5CDR - PubMed Abstract:
New antibacterials are needed to tackle antibiotic-resistant bacteria. Type IIA topoisomerases (topo2As), the targets of fluoroquinolones, regulate DNA topology by creating transient double-strand DNA breaks. Here we report the first co-crystal structures of the antibacterial QPT-1 and the anticancer drug etoposide with Staphylococcus aureus DNA gyrase, showing binding at the same sites in the cleaved DNA as the fluoroquinolone moxifloxacin. Unlike moxifloxacin, QPT-1 and etoposide interact with conserved GyrB TOPRIM residues rationalizing why QPT-1 can overcome fluoroquinolone resistance. Our data show etoposide's antibacterial activity is due to DNA gyrase inhibition and suggests other anticancer agents act similarly. Analysis of multiple DNA gyrase co-crystal structures, including asymmetric cleavage complexes, led to a 'pair of swing-doors' hypothesis in which the movement of one DNA segment regulates cleavage and religation of the second DNA duplex. This mechanism can explain QPT-1's bacterial specificity. Structure-based strategies for developing topo2A antibacterials are suggested.
Organizational Affiliation:
Antibacterial Discovery Performance Unit, Infectious Diseases, Therapy Area Unit, GlaxoSmithKline, 1250 South Collegeville Road, Collegeville, Pennsylvania 19426-0989, USA.