Trading off stability against activity in extremophilic aldolases.
Dick, M., Weiergraber, O.H., Classen, T., Bisterfeld, C., Bramski, J., Gohlke, H., Pietruszka, J.(2016) Sci Rep 6: 17908-17908
- PubMed: 26783049
- DOI: https://doi.org/10.1038/srep17908
- Primary Citation of Related Structures:
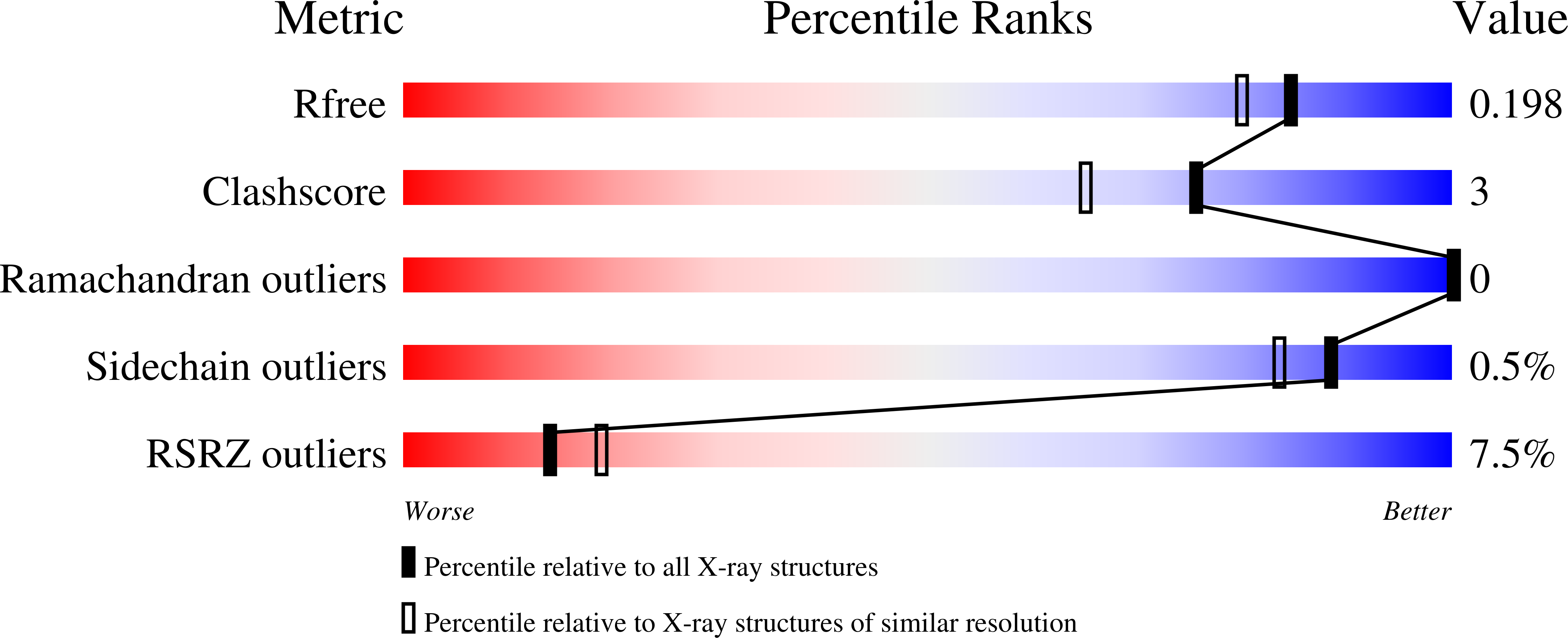
5C2X, 5C5Y, 5C6M - PubMed Abstract:
Understanding enzyme stability and activity in extremophilic organisms is of great biotechnological interest, but many questions are still unsolved. Using 2-deoxy-D-ribose-5-phosphate aldolase (DERA) as model enzyme, we have evaluated structural and functional characteristics of different orthologs from psychrophilic, mesophilic and hyperthermophilic organisms. We present the first crystal structures of psychrophilic DERAs, revealing a dimeric organization resembling their mesophilic but not their thermophilic counterparts. Conversion into monomeric proteins showed that the native dimer interface contributes to stability only in the hyperthermophilic enzymes. Nevertheless, introduction of a disulfide bridge in the interface of a psychrophilic DERA did confer increased thermostability, suggesting a strategy for rational design of more durable enzyme variants. Constraint network analysis revealed particularly sparse interactions between the substrate pocket and its surrounding α-helices in psychrophilic DERAs, which indicates that a more flexible active center underlies their high turnover numbers.
Organizational Affiliation:
Institute of Bioorganic Chemistry, Heinrich-Heine-Universität Düsseldorf im Forschungszentrum Jülich, and Bioeconomy Science Center (BioSC), Jülich, Germany.