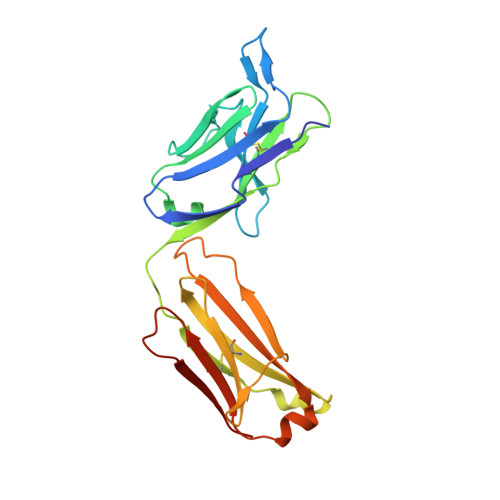
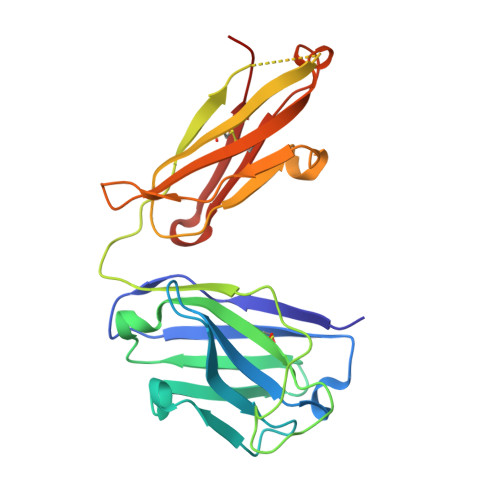
Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies.
Triller, G., Scally, S.W., Costa, G., Pissarev, M., Kreschel, C., Bosch, A., Marois, E., Sack, B.K., Murugan, R., Salman, A.M., Janse, C.J., Khan, S.M., Kappe, S.H.I., Adegnika, A.A., Mordmuller, B., Levashina, E.A., Julien, J.P., Wardemann, H.(2017) Immunity 47: 1197-1209.e10
- PubMed: 29195810
- DOI: https://doi.org/10.1016/j.immuni.2017.11.007
- Primary Citation of Related Structures:
5BK0, 5BK3, 5BK5, 6AZM, 6AZX - PubMed Abstract:
Antibodies against the NANP repeat of circumsporozoite protein (CSP), the major surface antigen of Plasmodium falciparum (Pf) sporozoites, can protect from malaria in animal models but protective humoral immunity is difficult to induce in humans. Here we cloned and characterized rare affinity-matured human NANP-reactive memory B cell antibodies elicited by natural Pf exposure that potently inhibited parasite transmission and development in vivo. We unveiled the molecular details of antibody binding to two distinct protective epitopes within the NANP repeat. NANP repeat recognition was largely mediated by germline encoded and immunoglobulin (Ig) heavy-chain complementarity determining region 3 (HCDR3) residues, whereas affinity maturation contributed predominantly to stabilizing the antigen-binding site conformation. Combined, our findings illustrate the power of exploring human anti-CSP antibody responses to develop tools for malaria control in the mammalian and the mosquito vector and provide a molecular basis for the structure-based design of next-generation CSP malaria vaccines.
Organizational Affiliation:
B Cell Immunology, German Cancer Research Center, Heidelberg, 69120, Germany.