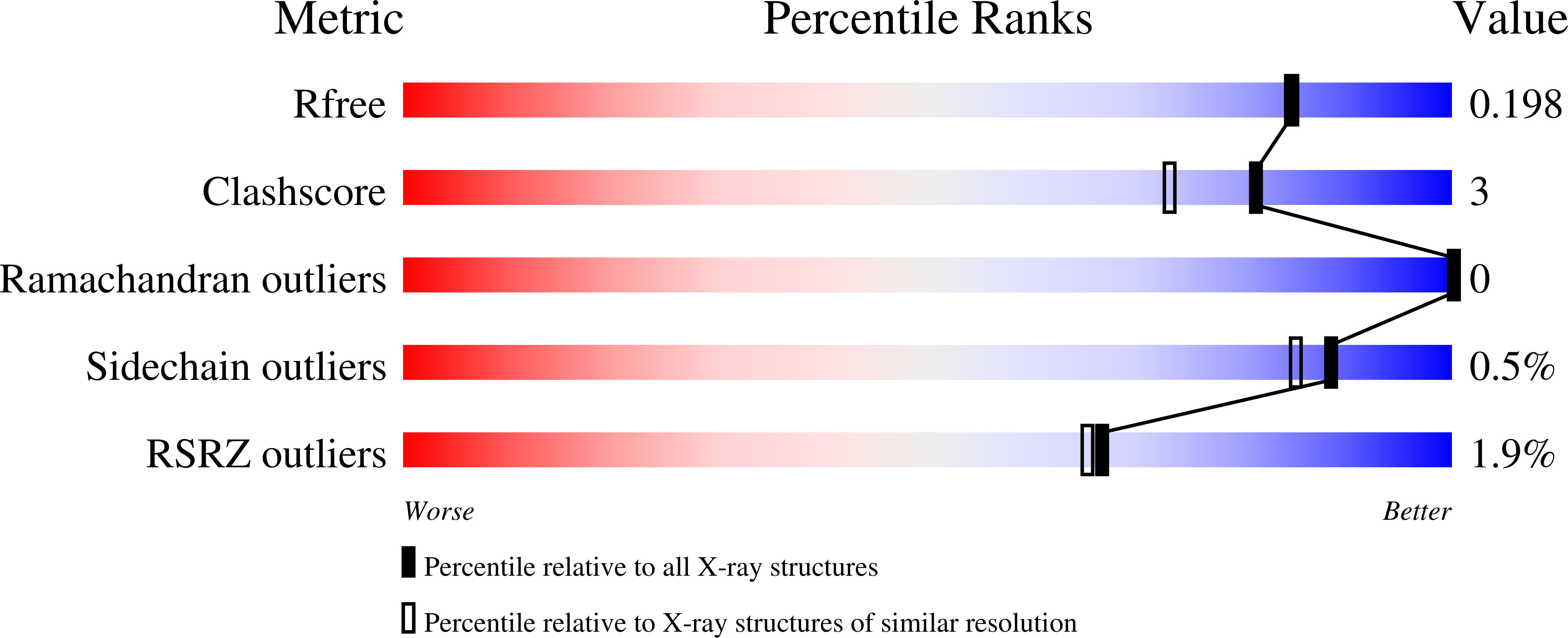
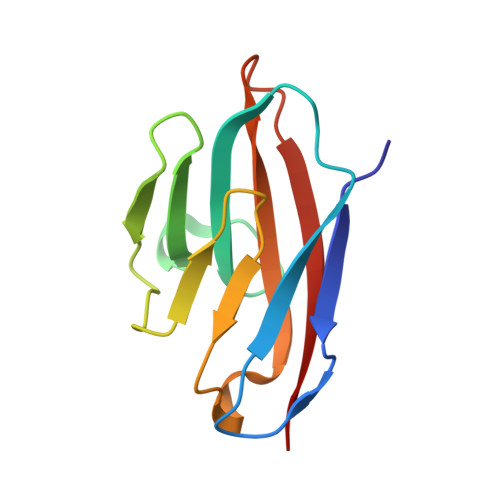
Diverse oligomeric states of CEACAM IgV domains.
Bonsor, D.A., Gunther, S., Beadenkopf, R., Beckett, D., Sundberg, E.J.(2015) Proc Natl Acad Sci U S A 112: 13561-13566
- PubMed: 26483485
- DOI: https://doi.org/10.1073/pnas.1509511112
- Primary Citation of Related Structures:
4Y88, 4Y8A, 4YIQ - PubMed Abstract:
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) comprise a large family of cell surface adhesion molecules that bind to themselves and other family members to carry out numerous cellular functions, including proliferation, signaling, differentiation, tumor suppression, and survival. They also play diverse and significant roles in immunity and infection. The formation of CEACAM oligomers is caused predominantly by interactions between their N-terminal IgV domains. Although X-ray crystal structures of CEACAM IgV domain homodimers have been described, how CEACAMs form heterodimers or remain monomers is poorly understood. To address this key aspect of CEACAM function, we determined the crystal structures of IgV domains that form a homodimeric CEACAM6 complex, monomeric CEACAM8, and a heterodimeric CEACAM6-CEACAM8 complex. To confirm and quantify these interactions in solution, we used analytical ultracentrifugation to measure the dimerization constants of CEACAM homodimers and isothermal titration calorimetry to determine the thermodynamic parameters and binding affinities of CEACAM heterodimers. We found the CEACAM6-CEACAM8 heterodimeric state to be substantially favored energetically relative to the CEACAM6 homodimer. Our data provide a molecular basis for the adoption of the diverse oligomeric states known to exist for CEACAMs and suggest ways in which CEACAM6 and CEACAM8 regulate the biological functions of one another, as well as of additional CEACAMs with which they interact, both in cis and in trans.
Organizational Affiliation:
Institute of Human Virology, University of Maryland School of Medicine, Baltimore, MD 21201;