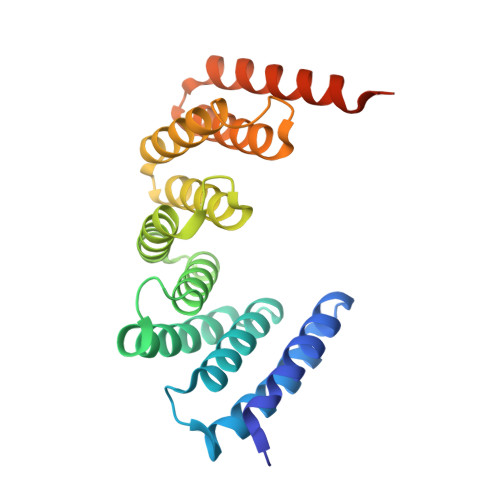
A Naturally Occurring Repeat Protein with High Internal Sequence Identity Defines a New Class of TPR-like Proteins.
Marold, J.D., Kavran, J.M., Bowman, G.D., Barrick, D.(2015) Structure 23: 2055-2065
- PubMed: 26439765
- DOI: https://doi.org/10.1016/j.str.2015.07.022
- Primary Citation of Related Structures:
4Y6C, 4Y6W - PubMed Abstract:
Linear repeat proteins often have high structural similarity and low (∼25%) pairwise sequence identities (PSI) among modules. We identified a unique P. anserina (Pa) sequence with tetratricopeptide repeat (TPR) homology, which contains longer (42 residue) repeats (42PRs) with an average PSI >91%. We determined the crystal structure of five tandem Pa 42PRs to 1.6 Å, and examined the stability and solution properties of constructs containing three to six Pa 42PRs. Compared with 34-residue TPRs (34PRs), Pa 42PRs have a one-turn extension of each helix, and bury more surface area. Unfolding transitions shift to higher denaturant concentration and become sharper as repeats are added. Fitted Ising models show Pa 42PRs to be more cooperative than consensus 34PRs, with increased magnitudes of intrinsic and interfacial free energies. These results demonstrate the tolerance of the TPR motif to length variation, and provide a basis to understand the effects of helix length on intrinsic/interfacial stability.
Organizational Affiliation:
T.C. Jenkins Department of Biophysics, Johns Hopkins University, Baltimore, MD 21218, USA.