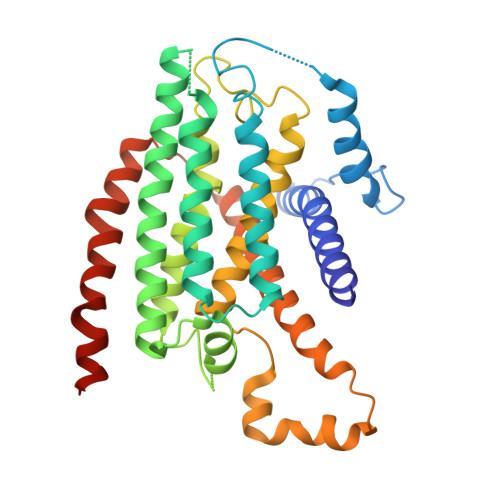
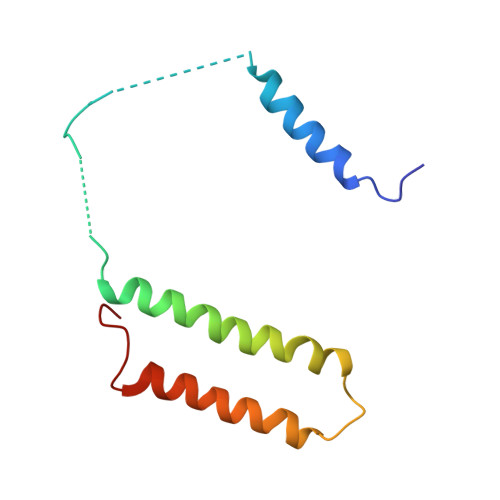
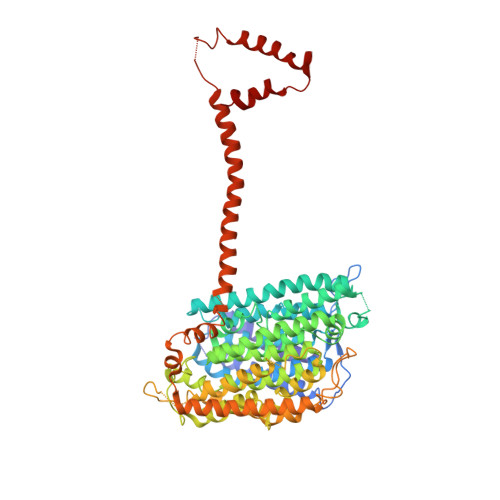
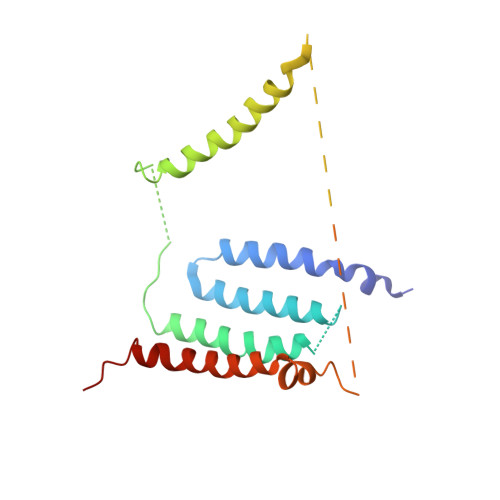
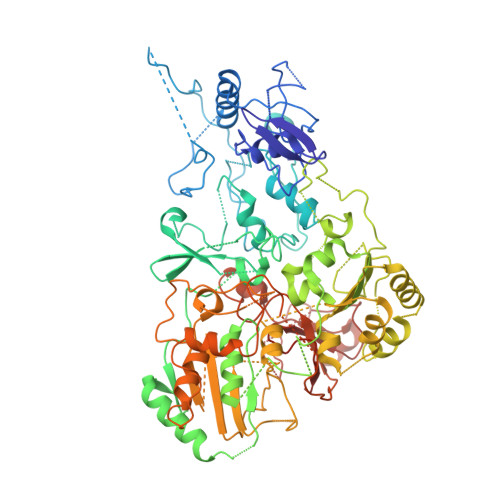
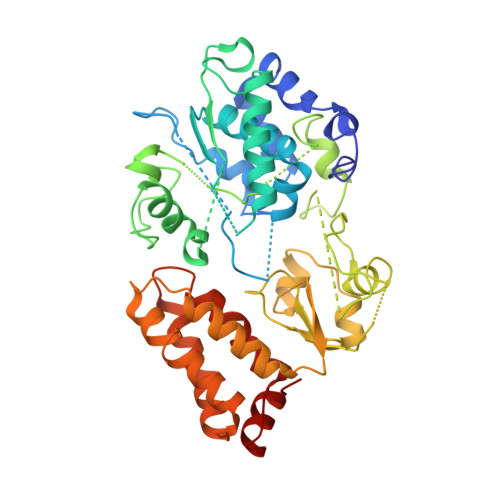
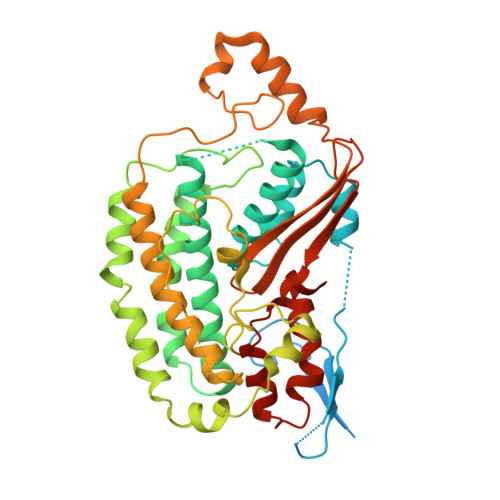
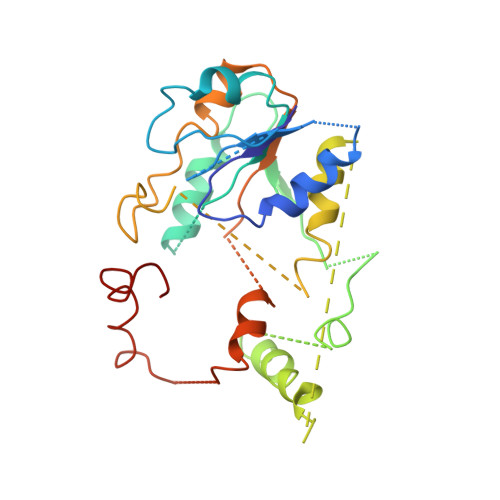
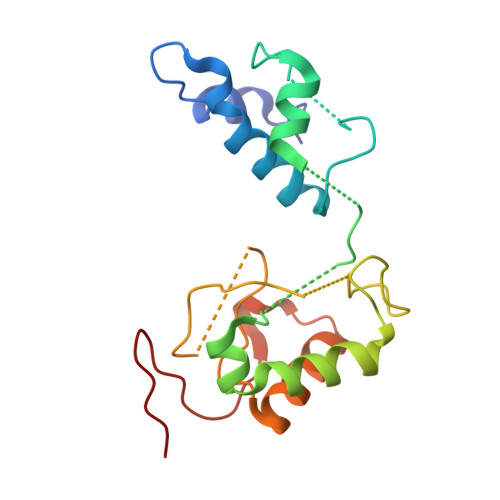
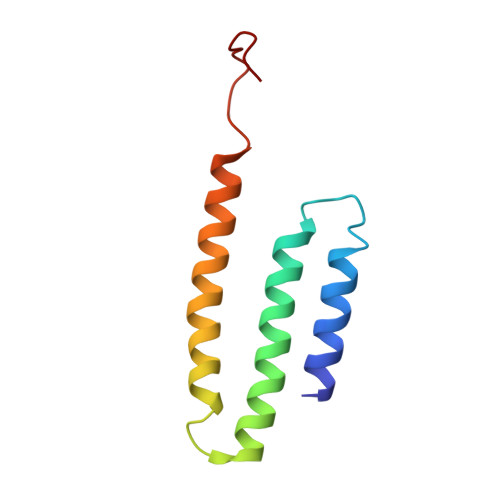
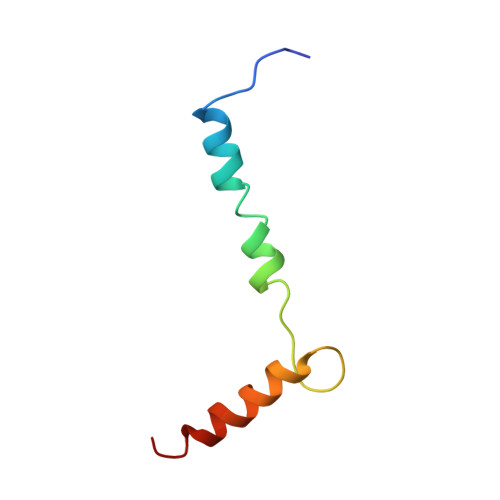
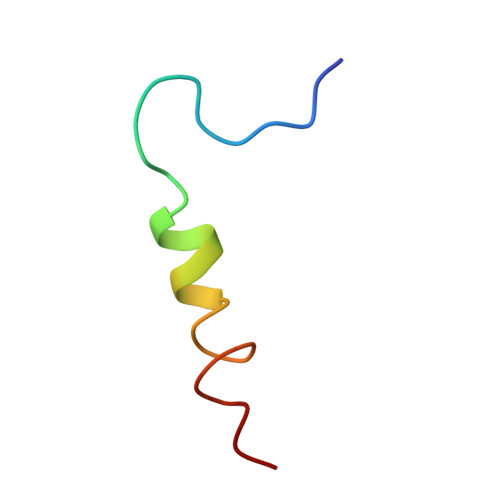
Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I.
Zickermann, V., Wirth, C., Nasiri, H., Siegmund, K., Schwalbe, H., Hunte, C., Brandt, U.(2015) Science 347: 44-49
- PubMed: 25554780
- DOI: https://doi.org/10.1126/science.1259859
- Primary Citation of Related Structures:
4WZ7 - PubMed Abstract:
Proton-pumping complex I of the mitochondrial respiratory chain is among the largest and most complicated membrane protein complexes. The enzyme contributes substantially to oxidative energy conversion in eukaryotic cells. Its malfunctions are implicated in many hereditary and degenerative disorders. We report the x-ray structure of mitochondrial complex I at a resolution of 3.6 to 3.9 angstroms, describing in detail the central subunits that execute the bioenergetic function. A continuous axis of basic and acidic residues running centrally through the membrane arm connects the ubiquinone reduction site in the hydrophilic arm to four putative proton-pumping units. The binding position for a substrate analogous inhibitor and blockage of the predicted ubiquinone binding site provide a model for the "deactive" form of the enzyme. The proposed transition into the active form is based on a concerted structural rearrangement at the ubiquinone reduction site, providing support for a two-state stabilization-change mechanism of proton pumping.
Organizational Affiliation:
Structural Bioenergetics Group, Institute of Biochemistry II, Medical School, Goethe-University, 60438 Frankfurt am Main, Germany. Cluster of Excellence Frankfurt "Macromolecular Complexes," Goethe-University, 60438 Frankfurt am Main, Germany. zickermann@med.uni-frankfurt.de carola.hunte@biochemie.uni-freiburg.de ulrich.brandt@radboudumc.nl.