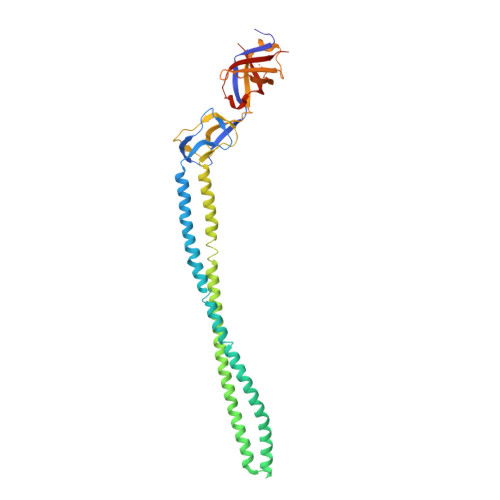
Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump.
Hinchliffe, P., Greene, N.P., Paterson, N.G., Crow, A., Hughes, C., Koronakis, V.(2014) FEBS Lett 588: 3147-3153
- PubMed: 24996185
- DOI: https://doi.org/10.1016/j.febslet.2014.06.055
- Primary Citation of Related Structures:
4TKO - PubMed Abstract:
Periplasmic adaptor proteins are key components of bacterial tripartite efflux pumps. The 2.85 Å resolution structure of an MFS (major facilitator superfamily) pump adaptor, Aquifex aeolicus EmrA, shows linearly arranged α-helical coiled-coil, lipoyl, and β-barrel domains, but lacks the fourth membrane-proximal domain shown in other pumps to interact with the inner membrane transporter. The adaptor α-hairpin, which binds outer membrane TolC, is exceptionally long at 127 Å, and the β-barrel contains a conserved disordered loop. The structure extends the view of adaptors as flexible, modular components that mediate diverse pump assembly, and suggests that in MFS tripartite pumps a hexamer of adaptors could provide a periplasmic seal.
Organizational Affiliation:
Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge CB2 1QP, UK.