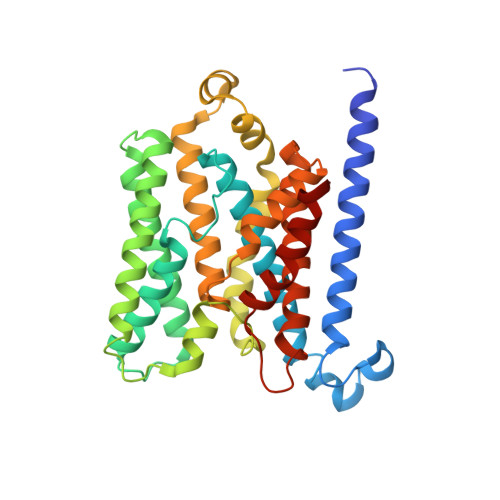
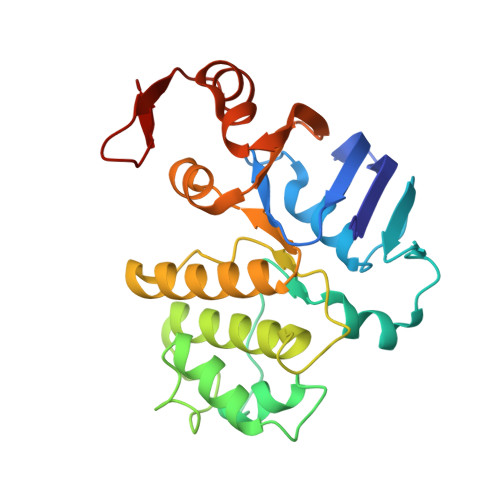
Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F.
Korkhov, V.M., Mireku, S.A., Veprintsev, D.B., Locher, K.P.(2014) Nat Struct Mol Biol 21: 1097-1099
- PubMed: 25402482
- DOI: https://doi.org/10.1038/nsmb.2918
- Primary Citation of Related Structures:
4R9U - PubMed Abstract:
The reaction mechanism of BtuCD-F-catalyzed vitamin B12 transport into Escherichia coli is currently unclear. Here we present the structure of the last missing state in the form of AMP-PNP-bound BtuCD, trapped by a disulfide cross-link. Our structural and biochemical data allow a consistent mechanism to be formulated, thus rationalizing the roles of substrate, ATP and substrate-binding protein.
Organizational Affiliation:
1] Institute of Molecular Biology and Biophysics, Eidgenössische Technische Hochschule Zurich, Zurich, Switzerland. [2] Laboratory of Biomolecular Research, Paul Scherrer Institute, Villigen, Switzerland.