A novel benzonitrile analogue inhibits rhinovirus replication.
Lacroix, C., Querol-Audi, J., Roche, M., Franco, D., Froeyen, M., Guerra, P., Terme, T., Vanelle, P., Verdaguer, N., Neyts, J., Leyssen, P.(2014) J Antimicrob Chemother 69: 2723-2732
- PubMed: 24948704
- DOI: https://doi.org/10.1093/jac/dku200
- Primary Citation of Related Structures:
4PDW - PubMed Abstract:
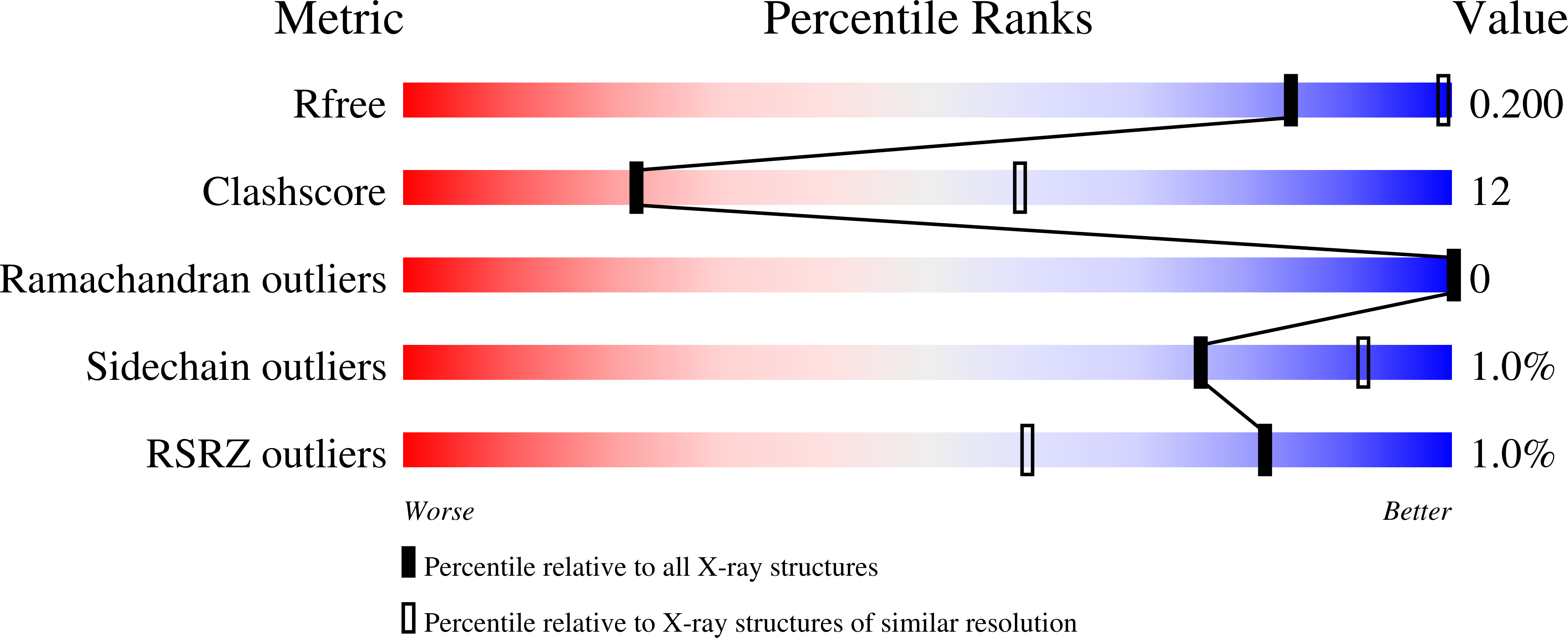
To study the characteristics and the mode of action of the anti-rhinovirus compound 4-[1-hydroxy-2-(4,5-dimethoxy-2-nitrophenyl)ethyl]benzonitrile (LPCRW_0005). The antiviral activity of LPCRW_0005 was evaluated in a cytopathic effect reduction assay against a panel of human rhinovirus (HRV) strains. To unravel its precise molecular mechanism of action, a time-of-drug-addition study, resistance selection and thermostability assays were performed. The crystal structure of the HRV14/LPCRW_0005 complex was elucidated as well. LPCRW_0005 proved to be a selective inhibitor of the replication of HRV14 (EC(50) of 2 ± 1 μM). Time-of-drug-addition studies revealed that LPCRW_0005 interferes with the earliest stages of virus replication. Phenotypic drug-resistant virus variants were obtained (≥30-fold decrease in susceptibility to the inhibitory effect of LPCRW_0005), which carried either an A150T or A150V amino acid substitution in the VP1 capsid protein. The link between the mutant genotype and drug-resistant phenotype was confirmed by reverse genetics. Cross-resistance studies and thermostability assays revealed that LPCRW_0005 has a similar mechanism of action to the capsid binder pleconaril. Elucidation of the crystal structure of the HRV14/LPCRW_0005 complex revealed the existence of multiple hydrophobic and polar interactions between the VP1 pocket and LPCRW_0005. LPCRW_0005 is a novel inhibitor of HRV14 replication that acts as a capsid binder. The compound has a chemical structure that is markedly smaller than that of other capsid binders. Structural studies show that LPCRW_0005, in contrast to pleconaril, leaves the toe end of the pocket in VP1 empty. This suggests that extended analogues of LPCRW_0005 that fill the full cavity could be more potent inhibitors of rhinovirus replication.
Organizational Affiliation:
Laboratory of Virology and Experimental Chemotherapy, Rega Institute for Medical Research, KU Leuven, Leuven, Belgium.