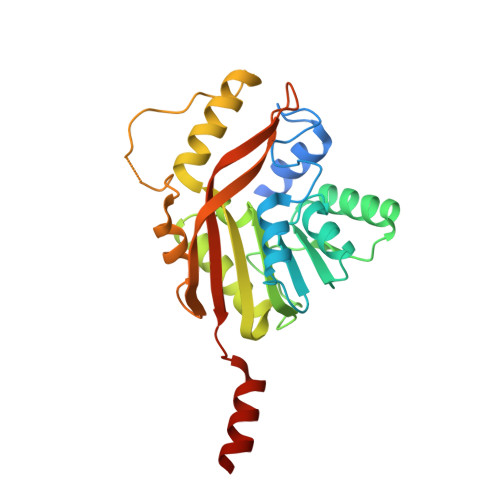
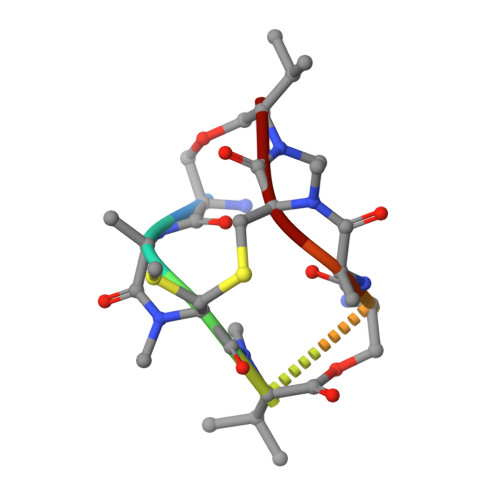
Conversion of a disulfide bond into a thioacetal group during echinomycin biosynthesis.
Hotta, K., Keegan, R.M., Ranganathan, S., Fang, M., Bibby, J., Winn, M.D., Sato, M., Lian, M., Watanabe, K., Rigden, D.J., Kim, C.Y.(2014) Angew Chem Int Ed Engl 53: 824-828
- PubMed: 24302672
- DOI: https://doi.org/10.1002/anie.201307404
- Primary Citation of Related Structures:
4NEC - PubMed Abstract:
Echinomycin is a nonribosomal depsipeptide natural product with a range of interesting bioactivities that make it an important target for drug discovery and development. It contains a thioacetal bridge, a unique chemical motif derived from the disulfide bond of its precursor antibiotic triostin A by the action of an S-adenosyl-L-methionine-dependent methyltransferase, Ecm18. The crystal structure of Ecm18 in complex with its reaction products S-adenosyl-L-homocysteine and echinomycin was determined at 1.50 Å resolution. Phasing was achieved using a new molecular replacement package called AMPLE, which automatically derives search models from structure predictions based on ab initio protein modelling. Structural analysis indicates that a combination of proximity effects, medium effects, and catalysis by strain drives the unique transformation of the disulfide bond into the thioacetal linkage.