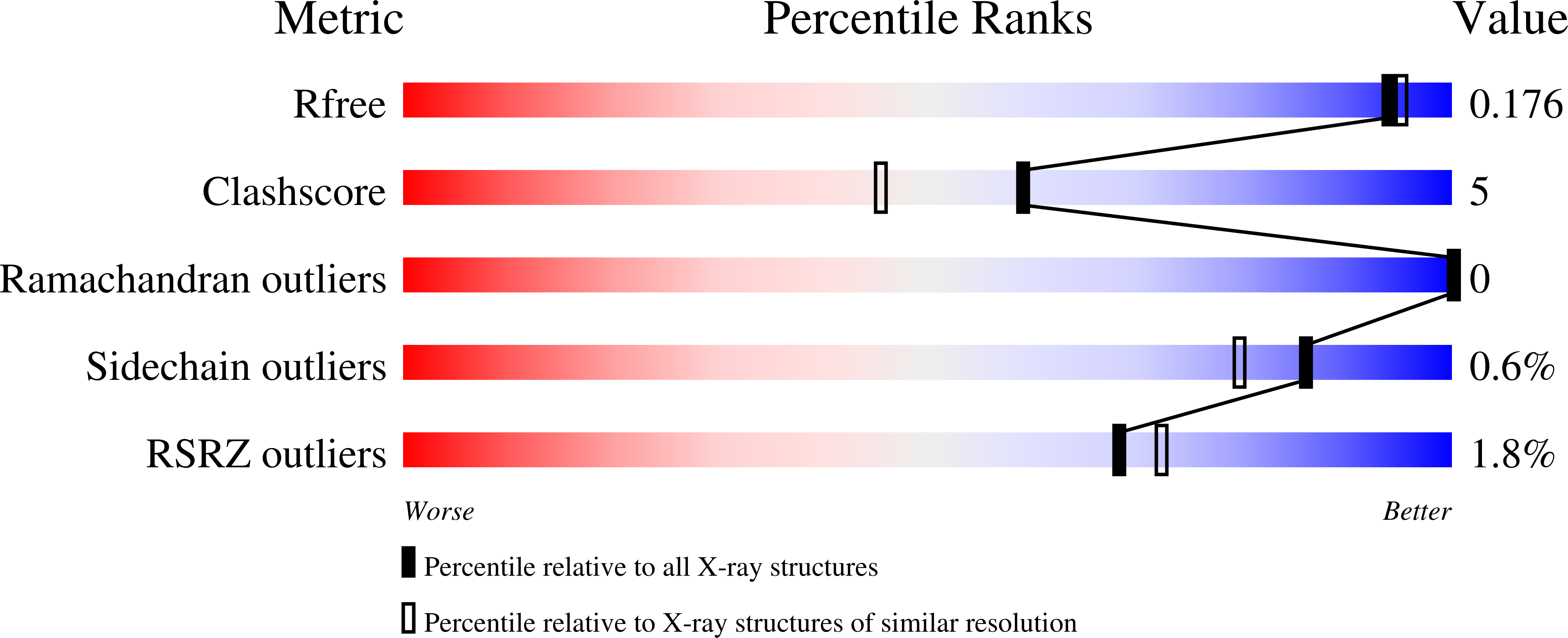
High-resolution structure of the N-terminal endonuclease domain of the lassa virus L polymerase in complex with magnesium ions.
Wallat, G.D., Huang, Q., Wang, W., Dong, H., Ly, H., Liang, Y., Dong, C.(2014) PLoS One 9: e87577-e87577
- PubMed: 24516554
- DOI: https://doi.org/10.1371/journal.pone.0087577
- Primary Citation of Related Structures:
4MIW - PubMed Abstract:
Lassa virus (LASV) causes deadly hemorrhagic fever disease for which there are no vaccines and limited treatments. LASV-encoded L polymerase is required for viral RNA replication and transcription. The functional domains of L-a large protein of 2218 amino acid residues-are largely undefined, except for the centrally located RNA-dependent RNA polymerase (RdRP) motif. Recent structural and functional analyses of the N-terminal region of the L protein from lymphocytic choriomeningitis virus (LCMV), which is in the same Arenaviridae family as LASV, have identified an endonuclease domain that presumably cleaves the cap structures of host mRNAs in order to initiate viral transcription. Here we present a high-resolution crystal structure of the N-terminal 173-aa region of the LASV L protein (LASV L173) in complex with magnesium ions at 1.72 Å. The structure is highly homologous to other known viral endonucleases of arena- (LCMV NL1), orthomyxo- (influenza virus PA), and bunyaviruses (La Crosse virus NL1). Although the catalytic residues (D89, E102 and K122) are highly conserved among the known viral endonucleases, LASV L endonuclease structure shows some notable differences. Our data collected from in vitro endonuclease assays and a reporter-based LASV minigenome transcriptional assay in mammalian cells confirm structural prediction of LASV L173 as an active endonuclease. The high-resolution structure of the LASV L endonuclease domain in complex with magnesium ions should aid the development of antivirals against lethal Lassa hemorrhagic fever.
Organizational Affiliation:
Biomedical Science Research Complex, School of Chemistry, University of St. Andrews, Fife, St. Andrews, United Kingdom.