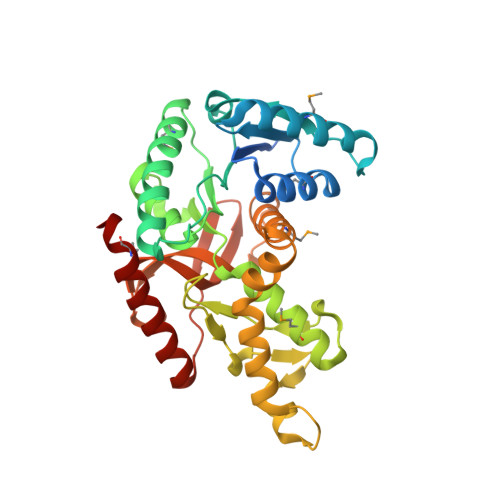
Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452
Malashkevich, V.N., Bonanno, J.B., Bhosle, R., Toro, R., Hillerich, B., Gizzi, A., Garforth, S., Kar, A., Chan, M.K., Lafluer, J., Patel, H., Matikainen, B., Chamala, S., Lim, S., Celikgil, A., Villegas, G., Evans, B., Love, J., Fiser, A., Khafizov, K., Seidel, R., Almo, S.C.To be published.