Structure-Efficiency Relationship of [1,2,4]Triazol-3-ylamines as Novel Nicotinamide Isosteres that Inhibit Tankyrases.
Shultz, M.D., Majumdar, D., Chin, D.N., Fortin, P.D., Feng, Y., Gould, T., Kirby, C.A., Stams, T., Waters, N.J., Shao, W.(2013) J Med Chem 56: 7049-7059
- PubMed: 23879431
- DOI: https://doi.org/10.1021/jm400826j
- Primary Citation of Related Structures:
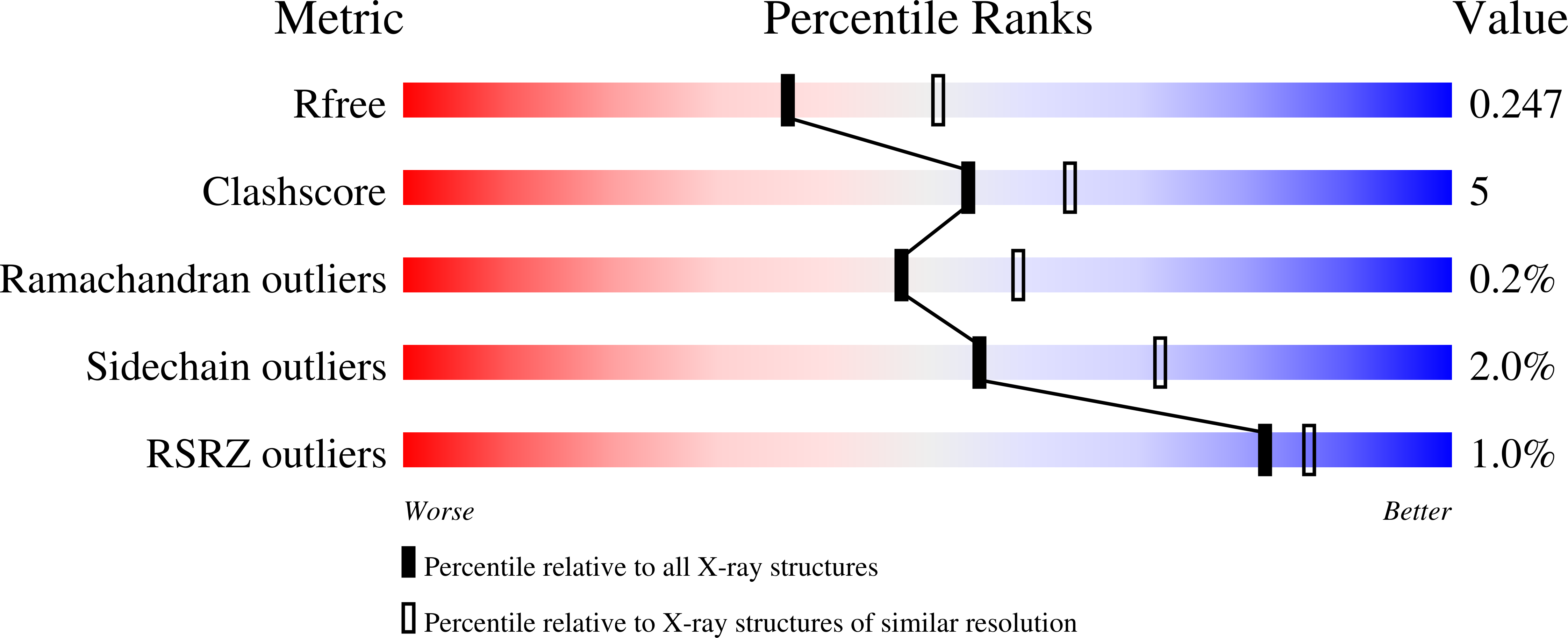
4KRS - PubMed Abstract:
Tankyrases 1 and 2 are members of the poly(ADP-ribose) polymerase (PARP) family of enzymes that modulate Wnt pathway signaling. While amide- and lactam-based nicotinamide mimetics that inhibit tankyrase activity, such as XAV939, are well-known, herein we report the discovery and evaluation of a novel nicotinamide isostere that demonstrates selectivity over other PARP family members. We demonstrate the utilization of lipophilic efficiency-based structure-efficiency relationships (SER) to rapidly drive the evaluation of this series. These efforts led to a series of selective, cell-active compounds with solubility, physicochemical, and in vitro properties suitable for further optimization.
Organizational Affiliation:
Novartis Institutes for Biomedical Research Incorporated , 250 Massachusetts Avenue, Cambridge, Massachusetts 02139, United States.