Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited.
Regan, M.C., Horanyi, P.S., Pryor, E.E., Sarver, J.L., Cafiso, D.S., Bushweller, J.H.(2013) Proc Natl Acad Sci U S A 110: 13374-13379
- PubMed: 23898196
- DOI: https://doi.org/10.1073/pnas.1301726110
- Primary Citation of Related Structures:
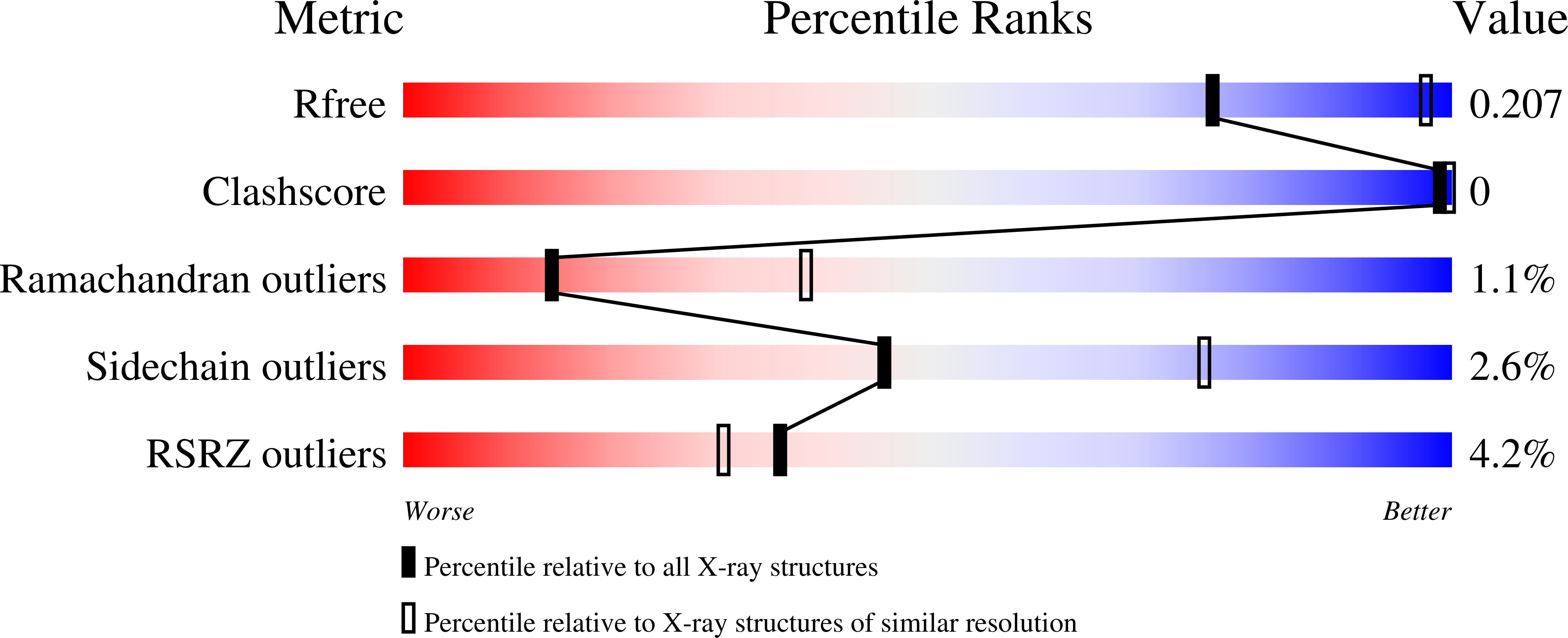
4IRG, 4IRH, 4IRI - PubMed Abstract:
The Ets-Related Gene (ERG) belongs to the Ets family of transcription factors and is critically important for maintenance of the hematopoietic stem cell population. A chromosomal translocation observed in the majority of human prostate cancers leads to the aberrant overexpression of ERG. We have identified regions flanking the ERG Ets domain responsible for autoinhibition of DNA binding and solved crystal structures of uninhibited, autoinhibited, and DNA-bound ERG. NMR-based measurements of backbone dynamics show that uninhibited ERG undergoes substantial dynamics on the millisecond-to-microsecond timescale but autoinhibited and DNA-bound ERG do not. We propose a mechanism whereby the allosteric basis of ERG autoinhibition is mediated predominantly by the regulation of Ets-domain dynamics with only modest structural changes.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908, USA.