Discovery of a class of novel tankyrase inhibitors that bind to both the nicotinamide pocket and the induced pocket.
Bregman, H., Gunaydin, H., Gu, Y., Schneider, S., Wilson, C., DiMauro, E.F., Huang, X.(2013) J Med Chem 56: 1341-1345
- PubMed: 23316926
- DOI: https://doi.org/10.1021/jm301607v
- Primary Citation of Related Structures:
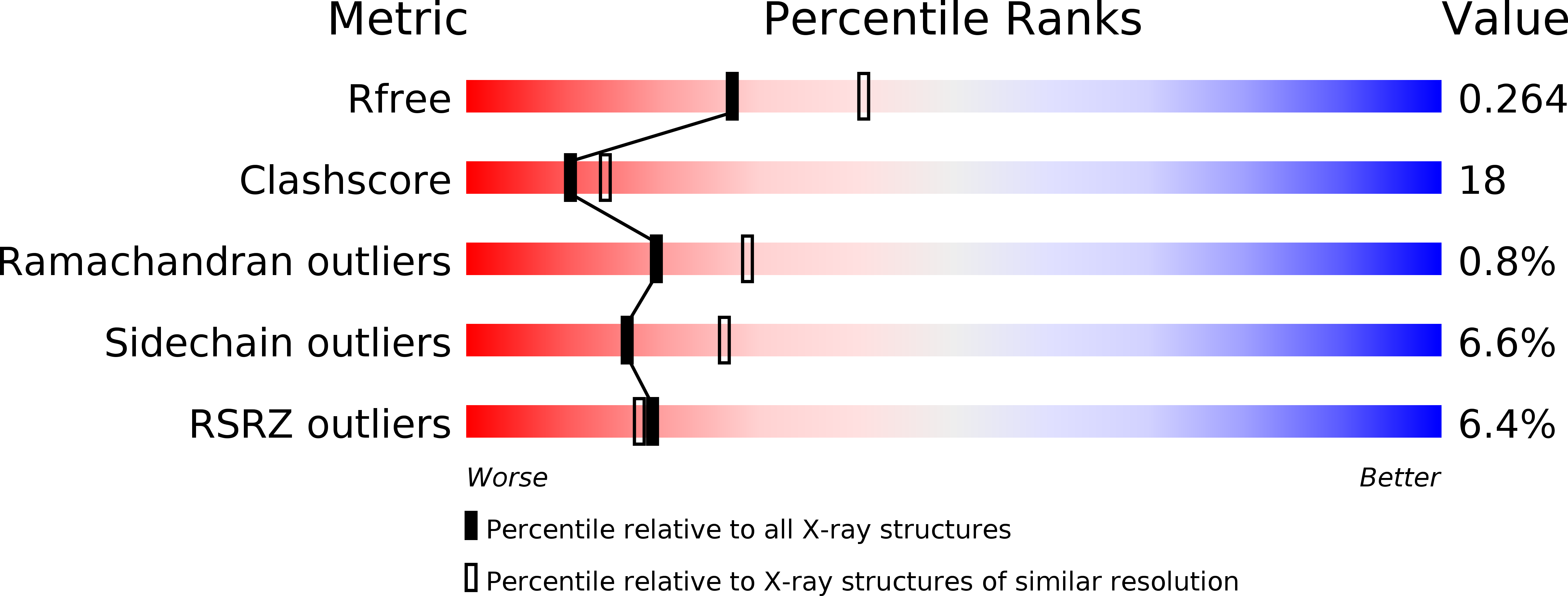
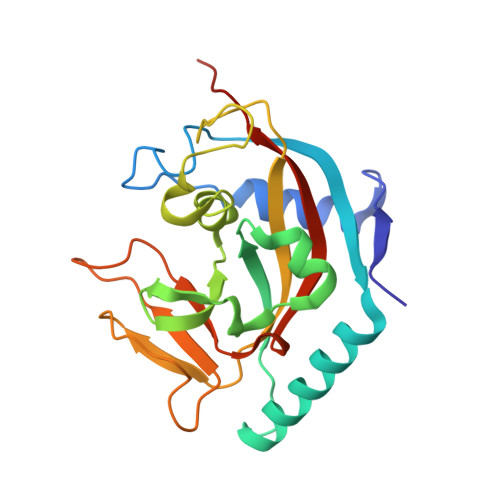
4I9I - PubMed Abstract:
Potent and selective inhibitors of tankyrases have recently been characterized to bind to an induced pocket. Here we report the identification of a novel potent and selective tankyrase inhibitor that binds to both the nicotinamide pocket and the induced pocket. The crystal structure of human TNKS1 in complex with this "dual-binder" provides a molecular basis for their strong and specific interactions and suggests clues for the further development of tankyrase inhibitors.
Organizational Affiliation:
Department of Medicinal Chemistry, Amgen Inc., 360 Binney Street, Cambridge, Massachusetts 02142, USA.